| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,356,704 – 2,356,805 |
| Length | 101 |
| Max. P | 0.683187 |

| Location | 2,356,704 – 2,356,805 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -21.86 |
| Energy contribution | -21.08 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
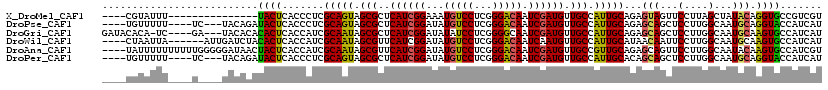
>X_DroMel_CAF1 2356704 101 + 22224390 ----CGUAUUU---------------UACUCACCCUCGCAGUAGCGCUCAUCGGAAAUGUCCUCGGGACAAUCGAUGUUGCCAUUGCAGAGUAGUUCCUUAGCUAUACAGGUGCCGUCGU ----.......---------------....((((((((((((.(((..((((((...(((((...))))).)))))).))).))))).)))((((......))))....))))....... ( -28.70) >DroPse_CAF1 1570 109 + 1 ----UGUUUUU----UC---UACAGAUACUCACCCUCGCAGUAGCGCUCAUCGGAUAUGUCCUCGGGACAAUCGAUGUUGCCAUUGCAGAGCAGCUCCUUGGCAAUGCAGGUACCAUCAU ----.......----..---....(((....(((...(((((.(((..((((((...(((((...))))).)))))).))).)))))...((((((....)))..))).)))...))).. ( -28.00) >DroGri_CAF1 1752 112 + 1 GAUACACA-UC----GA---UACACACACUCACCAUCGCAAUAGCGCUCAUCGGAUAUAUCCUCGGGGCAAUCGAUGUUGCCAUUGCAGAGCAGCUCCUUGGCAAUGCAAGUGCCAUCAU (((.((((-((----((---(................((....))((((...(((....)))...)))).))))))(((((((..((......))....)))))))....)))..))).. ( -30.70) >DroWil_CAF1 2754 110 + 1 ----CUAAUUA------AUUGAUCUACACUCACCAUCGCAAUAGCGUUCAUCGGAUAUGUCCUCGGGACAAUCAAUGUUGCCAUUGCAUAACAAUUCCUUGGCAAUGCAAGUGCCAUCAU ----.......------..(((...............(((((.(((..(((..(((.(((((...)))))))).))).))).)))))............(((((.......)))))))). ( -23.10) >DroAna_CAF1 1412 116 + 1 ----UAUUUUUUUUUUGGGGGAUAACUACUCACCAUCGCAAUAGCGUUCAUCGGAUAUGUCCUCGGGACAAUCGAUGUUGCCGUUGCAGAGCAGUUCCUUGGCAAUACAAGUGCCAUCGU ----.............(((((..(((.(((......((((..(((..((((((...(((((...))))).)))))).)))..)))).))).))))))))((((.......))))..... ( -32.40) >DroPer_CAF1 1560 109 + 1 ----UGUUUUU----UC---UACAGAUACUCACCCUCGCAGUAGCGCUCAUCGGAUAUGUCCUCGGGACAAUCGAUGUUGCCAUUGCACAGCAGCUCCUUGGCAAUGCAGGUACCAUCAU ----.......----..---....(((....(((...(((((.(((..((((((...(((((...))))).)))))).))).)))))...((((((....)))..))).)))...))).. ( -28.00) >consensus ____CGUAUUU____U____UACAGAUACUCACCAUCGCAAUAGCGCUCAUCGGAUAUGUCCUCGGGACAAUCGAUGUUGCCAUUGCAGAGCAGCUCCUUGGCAAUGCAAGUGCCAUCAU ..........................((((.......(((((.(((..((((((...(((((...))))).)))))).))).)))))...(((...(....)...))).))))....... (-21.86 = -21.08 + -0.78)

| Location | 2,356,704 – 2,356,805 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -23.45 |
| Energy contribution | -22.53 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
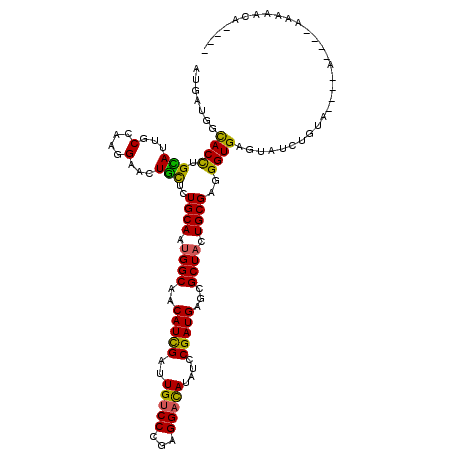
>X_DroMel_CAF1 2356704 101 - 22224390 ACGACGGCACCUGUAUAGCUAAGGAACUACUCUGCAAUGGCAACAUCGAUUGUCCCGAGGACAUUUCCGAUGAGCGCUACUGCGAGGGUGAGUA---------------AAAUACG---- ...((..((((....(((........)))(((.(((.((((..(((((..(((((...)))))....)))))...)))).)))))))))).)).---------------.......---- ( -28.00) >DroPse_CAF1 1570 109 - 1 AUGAUGGUACCUGCAUUGCCAAGGAGCUGCUCUGCAAUGGCAACAUCGAUUGUCCCGAGGACAUAUCCGAUGAGCGCUACUGCGAGGGUGAGUAUCUGUA---GA----AAAAACA---- ..((((.(((((((.((((((.((((...))))....))))))(((((..(((((...)))))....))))).))((....))..)))))..))))....---..----.......---- ( -32.40) >DroGri_CAF1 1752 112 - 1 AUGAUGGCACUUGCAUUGCCAAGGAGCUGCUCUGCAAUGGCAACAUCGAUUGCCCCGAGGAUAUAUCCGAUGAGCGCUAUUGCGAUGGUGAGUGUGUGUA---UC----GA-UGUGUAUC .((((((((((..((((((.((..(((.((((......(((((......)))))....(((....)))...))))))).))))))).)..)))))...))---))----).-........ ( -34.00) >DroWil_CAF1 2754 110 - 1 AUGAUGGCACUUGCAUUGCCAAGGAAUUGUUAUGCAAUGGCAACAUUGAUUGUCCCGAGGACAUAUCCGAUGAACGCUAUUGCGAUGGUGAGUGUAGAUCAAU------UAAUUAG---- .((((.(((((..(.....(((....)))...(((((((((..(((((..(((((...)))))....)))))...)))))))))...)..)))))..))))..------.......---- ( -34.10) >DroAna_CAF1 1412 116 - 1 ACGAUGGCACUUGUAUUGCCAAGGAACUGCUCUGCAACGGCAACAUCGAUUGUCCCGAGGACAUAUCCGAUGAACGCUAUUGCGAUGGUGAGUAGUUAUCCCCCAAAAAAAAAAUA---- ....(((((.......))))).(((((((((((((((..((..(((((..(((((...)))))....)))))...))..))))).....)))))))..)))...............---- ( -35.60) >DroPer_CAF1 1560 109 - 1 AUGAUGGUACCUGCAUUGCCAAGGAGCUGCUGUGCAAUGGCAACAUCGAUUGUCCCGAGGACAUAUCCGAUGAGCGCUACUGCGAGGGUGAGUAUCUGUA---GA----AAAAACA---- ..((((.(((((((.((((((..(.((....)).)..))))))(((((..(((((...)))))....))))).))((....))..)))))..))))....---..----.......---- ( -31.90) >consensus AUGAUGGCACCUGCAUUGCCAAGGAACUGCUCUGCAAUGGCAACAUCGAUUGUCCCGAGGACAUAUCCGAUGAGCGCUACUGCGAGGGUGAGUAUCUGUA____A____AAAAACA____ .......((((.(((...(....)...)))..((((.((((..(((((..(((((...)))))....)))))...)))).))))..)))).............................. (-23.45 = -22.53 + -0.91)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:02 2006