
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,849,901 – 20,850,001 |
| Length | 100 |
| Max. P | 0.910362 |

| Location | 20,849,901 – 20,850,001 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.78 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -6.78 |
| Energy contribution | -7.86 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.21 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20849901 100 + 22224390 UAUCGAAUUUGGUGGUAUGUGCCGUG------CUACA---AGCUCGUAU-------UUCGAAUUCCCGUGAAAUGAGCAGGAACAGCAGGCCGGGAUCAUCGCCU----ACAUAAGUUUA ..........((((((.((.(((.((------((.(.---.((((((.(-------(.((......)).)).))))))..)...)))))))))..))))))....----........... ( -25.20) >DroSec_CAF1 4909 104 + 1 UAUCGAAUUUGG---UAAGUGCCGUG------CUACGAAGAGCUCGUAU-------UUCGAGUUCCCCUGAAACGAGCAGGAACAGCUGGCCGGCCUCACCGCCUACCUACAUAAGUUUA ..........((---(.((.((((.(------((((...(((((((...-------..))))))).((((.......))))....).)))))))))).)))................... ( -31.50) >DroSim_CAF1 5925 104 + 1 UAUCGAAUUUGG---UAGGUGCCGUG------CUACGAAGAGCUCGUGU-------UUCGAGUUCCCCUGAAACGAGCAGGAGCAGCUGGCCGGCCUCACCGCCUACCUACAUAAGUUUA ....((((((((---(((((((((.(------((.(...(((((((...-------..))))))).((((.......)))).).))))))).(((......))).)))))).))))))). ( -38.00) >DroEre_CAF1 6066 92 + 1 UAUCGAAUUUGG---UAUGUGCCCAG------CUACC---AGCUCGGAU-------UUCGAAUUCCCCGAAAAUGAGCAGGAACAGCUGGCC---------AACUCACCACCUAAGUUUA .........(((---(..((..((((------((.((---.(((((..(-------((((.......))))).))))).))...))))))..---------.))..)))).......... ( -27.70) >DroYak_CAF1 5675 113 + 1 UAUCGAAUUUGG---UAUGUGUCCAGCUGCAGCUACG---AGUUCGGAUUCGGGAAUUCGAAUUCCCCGAAAACAAGCAGGAACAGCUGGCCA-CCUUACCAACUCACCACCUAAGUUUA ....((..((((---((.(((.(((((((.......(---((((((((((....))))))))))).((...........))..))))))).))-)..)))))).)).............. ( -35.60) >consensus UAUCGAAUUUGG___UAUGUGCCGUG______CUACG___AGCUCGUAU_______UUCGAAUUCCCCUGAAACGAGCAGGAACAGCUGGCCGGCCUCACCGCCUAACCACAUAAGUUUA ....(((((((.......(.(((.................((((((............))))))...........(((.......)))))))((........))........))))))). ( -6.78 = -7.86 + 1.08)



| Location | 20,849,901 – 20,850,001 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.78 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -7.94 |
| Energy contribution | -8.10 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.23 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20849901 100 - 22224390 UAAACUUAUGU----AGGCGAUGAUCCCGGCCUGCUGUUCCUGCUCAUUUCACGGGAAUUCGAA-------AUACGAGCU---UGUAG------CACGGCACAUACCACCAAAUUCGAUA .........((----((((..........)))))).(((((((.........)))))))(((((-------......((.---....)------)..((......))......))))).. ( -21.40) >DroSec_CAF1 4909 104 - 1 UAAACUUAUGUAGGUAGGCGGUGAGGCCGGCCAGCUGUUCCUGCUCGUUUCAGGGGAACUCGAA-------AUACGAGCUCUUCGUAG------CACGGCACUUA---CCAAAUUCGAUA ....(((((....))))).((((((((((....((((.....(((((((((..(....)..)))-------..))))))......)))------).)))).))))---)).......... ( -34.10) >DroSim_CAF1 5925 104 - 1 UAAACUUAUGUAGGUAGGCGGUGAGGCCGGCCAGCUGCUCCUGCUCGUUUCAGGGGAACUCGAA-------ACACGAGCUCUUCGUAG------CACGGCACCUA---CCAAAUUCGAUA ............(((((((((.....)))(((.(((((....(((((((((..(....)..)))-------..)))))).....))))------)..))).))))---)).......... ( -38.60) >DroEre_CAF1 6066 92 - 1 UAAACUUAGGUGGUGAGUU---------GGCCAGCUGUUCCUGCUCAUUUUCGGGGAAUUCGAA-------AUCCGAGCU---GGUAG------CUGGGCACAUA---CCAAAUUCGAUA ..........(((((.((.---------(.(((((((..((.((((..((((((.....)))))-------)...)))).---)))))------)))).))).))---)))......... ( -32.30) >DroYak_CAF1 5675 113 - 1 UAAACUUAGGUGGUGAGUUGGUAAGG-UGGCCAGCUGUUCCUGCUUGUUUUCGGGGAAUUCGAAUUCCCGAAUCCGAACU---CGUAGCUGCAGCUGGACACAUA---CCAAAUUCGAUA .............((((((((((..(-((.((((((((..(((((((..((((((((.......))))))))..)))...---.))))..)))))))).))).))---)).))))))... ( -44.30) >consensus UAAACUUAUGUAGGGAGGCGGUGAGGCCGGCCAGCUGUUCCUGCUCGUUUCAGGGGAAUUCGAA_______AUACGAGCU___CGUAG______CACGGCACAUA___CCAAAUUCGAUA .............................(((....((((((...........))))))(((............)))....................))).................... ( -7.94 = -8.10 + 0.16)

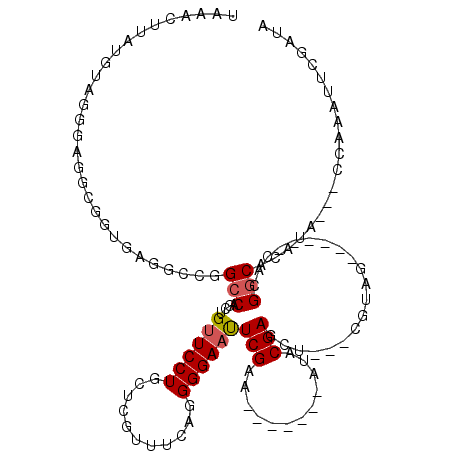
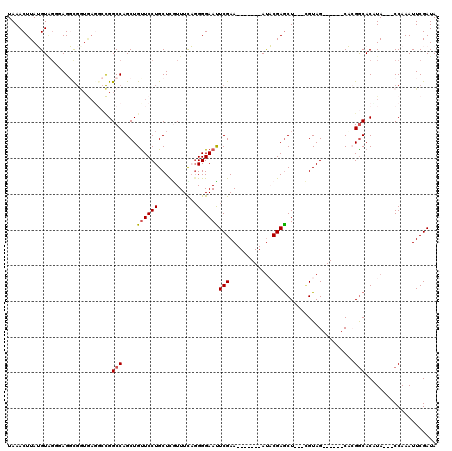
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:57 2006