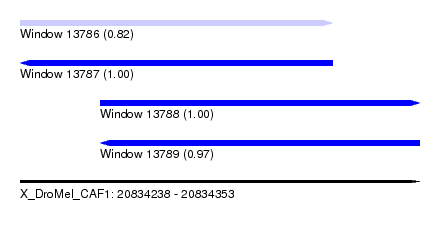
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,834,238 – 20,834,353 |
| Length | 115 |
| Max. P | 0.999961 |

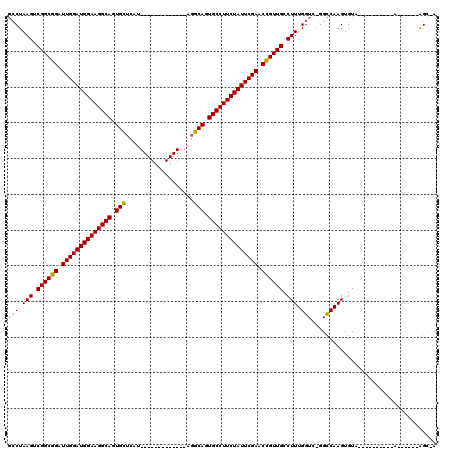
| Location | 20,834,238 – 20,834,328 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.66 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -15.14 |
| Energy contribution | -16.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20834238 90 + 22224390 UGGCU-----------------GACACUUGGCCCGACCAAAGGCAACAGUUCGAAUAGAAGGCACUGCCU-------------AUGUGCACUGCCUUCCAUCCAAUCCGCCGACUUAGGC (((.(-----------------(..(((..(((........)))...))).......(((((((.(((..-------------....))).))))))))).)))....(((......))) ( -26.10) >DroSim_CAF1 6671 102 + 1 --GCCACACAAUCCGCACACAAUACACUUGGCC--ACCAAAGGCAACGGUUCGAAUAGAAGGCACUGCC--------------AUGAGCACUGCCUUCCAUCCAAUCCGCCGACUUAGGC --(((.........................(((--......)))..((((..((((.(((((((.(((.--------------....))).))))))).))....)).)))).....))) ( -26.80) >DroYak_CAF1 797 107 + 1 UAACU---UGGU----------AACACUUGACCUGACCAAAGCCAACGGUUCGAAUAGAAGGCACUGCCUAUAGGCGACGCCUAUAAACACUGCCUUCCAUCCAAUCCGCCGACUUAGUG (((.(---((((----------.....((((.(((...........))).))))...(((((((.((..(((((((...)))))))..)).)))))))..........))))).)))... ( -28.30) >consensus U_GCU______U__________AACACUUGGCC_GACCAAAGGCAACGGUUCGAAUAGAAGGCACUGCCU_____________AUGAGCACUGCCUUCCAUCCAAUCCGCCGACUUAGGC ...........................(((((.........(....)((((.((...(((((((.(((...................))).)))))))..)).)))).)))))....... (-15.14 = -16.14 + 1.00)



| Location | 20,834,238 – 20,834,328 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.66 |
| Mean single sequence MFE | -48.43 |
| Consensus MFE | -36.52 |
| Energy contribution | -36.74 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -5.34 |
| Structure conservation index | 0.75 |
| SVM decision value | 4.90 |
| SVM RNA-class probability | 0.999961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20834238 90 - 22224390 GCCUAAGUCGGCGGAUUGGAUGGAAGGCAGUGCACAU-------------AGGCAGUGCCUUCUAUUCGAACUGUUGCCUUUGGUCGGGCCAAGUGUC-----------------AGCCA (((.(((.((((((.((((((((((((((.(((....-------------..))).)))))))))))))).)))))).))).)))..(((........-----------------.))). ( -42.50) >DroSim_CAF1 6671 102 - 1 GCCUAAGUCGGCGGAUUGGAUGGAAGGCAGUGCUCAU--------------GGCAGUGCCUUCUAUUCGAACCGUUGCCUUUGGU--GGCCAAGUGUAUUGUGUGCGGAUUGUGUGGC-- (((.(((.((((((.((((((((((((((.(((....--------------.))).)))))))))))))).)))))).))).)))--.((((......(((....)))......))))-- ( -46.80) >DroYak_CAF1 797 107 - 1 CACUAAGUCGGCGGAUUGGAUGGAAGGCAGUGUUUAUAGGCGUCGCCUAUAGGCAGUGCCUUCUAUUCGAACCGUUGGCUUUGGUCAGGUCAAGUGUU----------ACCA---AGUUA .(((((((((((((.((((((((((((((.(((((((((((...))))))))))).)))))))))))))).)))))))).)))))..(((........----------))).---..... ( -56.00) >consensus GCCUAAGUCGGCGGAUUGGAUGGAAGGCAGUGCUCAU_____________AGGCAGUGCCUUCUAUUCGAACCGUUGCCUUUGGUC_GGCCAAGUGUA__________A______AGC_A (((.(((.((((((.((((((((((((((.(((...................))).)))))))))))))).)))))).))).)))................................... (-36.52 = -36.74 + 0.23)


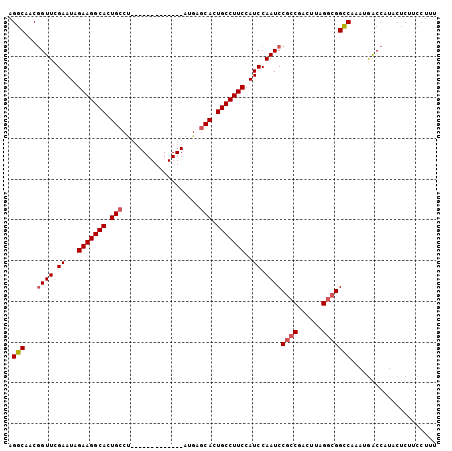
| Location | 20,834,261 – 20,834,353 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.31 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20834261 92 + 22224390 AGGCAACAGUUCGAAUAGAAGGCACUGCCU-------------AUGUGCACUGCCUUCCAUCCAAUCCGCCGACUUAGGCGGCCAAAUGACCAUACUCUUCCUUU .(((.............(((((((.(((..-------------....))).))))))).........((((......)))))))..................... ( -27.60) >DroSim_CAF1 6707 91 + 1 AGGCAACGGUUCGAAUAGAAGGCACUGCC--------------AUGAGCACUGCCUUCCAUCCAAUCCGCCGACUUAGGCGGCCAAAUGGCCAUACUCGUCCUUC .(....)((..(((...(((((((.(((.--------------....))).)))))))..........(((......)))((((....))))....))).))... ( -33.10) >DroYak_CAF1 824 105 + 1 AGCCAACGGUUCGAAUAGAAGGCACUGCCUAUAGGCGACGCCUAUAAACACUGCCUUCCAUCCAAUCCGCCGACUUAGUGGGGCCCCUAACCAUCCACUUCCUUU ......((((..((((.(((((((.((..(((((((...)))))))..)).))))))).))....)).))))....((((((...........))))))...... ( -29.90) >consensus AGGCAACGGUUCGAAUAGAAGGCACUGCCU_____________AUGAGCACUGCCUUCCAUCCAAUCCGCCGACUUAGGCGGCCAAAUGACCAUACUCUUCCUUU .(((...((((.((...(((((((.(((...................))).)))))))..)).))))((((......)))))))..................... (-21.42 = -22.31 + 0.89)


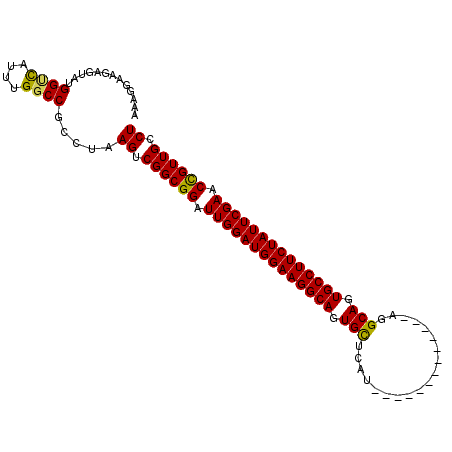
| Location | 20,834,261 – 20,834,353 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -49.07 |
| Consensus MFE | -37.85 |
| Energy contribution | -37.41 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -6.14 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20834261 92 - 22224390 AAAGGAAGAGUAUGGUCAUUUGGCCGCCUAAGUCGGCGGAUUGGAUGGAAGGCAGUGCACAU-------------AGGCAGUGCCUUCUAUUCGAACUGUUGCCU ..(((((.(((............(((((......))))).((((((((((((((.(((....-------------..))).))))))))))))))))).)).))) ( -41.40) >DroSim_CAF1 6707 91 - 1 GAAGGACGAGUAUGGCCAUUUGGCCGCCUAAGUCGGCGGAUUGGAUGGAAGGCAGUGCUCAU--------------GGCAGUGCCUUCUAUUCGAACCGUUGCCU ....(((.....(((((....))))).....)))(((((.((((((((((((((.(((....--------------.))).)))))))))))))).))))).... ( -45.90) >DroYak_CAF1 824 105 - 1 AAAGGAAGUGGAUGGUUAGGGGCCCCACUAAGUCGGCGGAUUGGAUGGAAGGCAGUGUUUAUAGGCGUCGCCUAUAGGCAGUGCCUUCUAUUCGAACCGUUGGCU ......(((((..(((.....)))))))).(((((((((.((((((((((((((.(((((((((((...))))))))))).)))))))))))))).))))))))) ( -59.90) >consensus AAAGGAAGAGUAUGGUCAUUUGGCCGCCUAAGUCGGCGGAUUGGAUGGAAGGCAGUGCUCAU_____________AGGCAGUGCCUUCUAUUCGAACCGUUGCCU .............((((....)))).....((.((((((.((((((((((((((.(((...................))).)))))))))))))).)))))).)) (-37.85 = -37.41 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:55 2006