
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,811,741 – 20,811,875 |
| Length | 134 |
| Max. P | 0.664235 |

| Location | 20,811,741 – 20,811,835 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.57 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20811741 94 + 22224390 --------------------------GUGUGGGACCACAUGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCAACGG --------------------------.(((((..(((...(((....)))....)))))))).((((((((((((((((.....)))))))..))))))))).(((((.....)).))). ( -22.40) >DroEre_CAF1 3800 120 + 1 GUGGGGGGAUGGGAGGGGGGUGUGGGGUGUGGGACCACAUGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCAACGG ((((.((((..(((((.(.((((((.........)))))).).)))))...))))..))))..((((((((((((((((.....)))))))..))))))))).(((((.....)).))). ( -38.10) >DroYak_CAF1 8042 97 + 1 GUG-----------------------GUGUGGGACCACAUGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCAACGG (((-----------------------((..((((....(((......)))))))..)))))..((((((((((((((((.....)))))))..))))))))).(((((.....)).))). ( -24.70) >consensus GUG_______________________GUGUGGGACCACAUGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCAACGG ...........................(((((..(((...(((....)))....)))))))).((((((((((((((((.....)))))))..))))))))).(((((.....)).))). (-22.40 = -22.40 + 0.00)



| Location | 20,811,741 – 20,811,835 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.57 |
| Mean single sequence MFE | -14.60 |
| Consensus MFE | -14.30 |
| Energy contribution | -14.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.16 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20811741 94 - 22224390 CCGUUGCUUGUUACACGUCUAAUCUGUUAAAUCCUGUUUACCAAGAUUACAAAUCAAUUGUGGCCAGGAAAUAAAAACACAUGUGGUCCCACAC-------------------------- ..(((..((((((((.(......))))....(((((..(((...(((.....)))....)))..)))))))))).)))...((((....)))).-------------------------- ( -14.30) >DroEre_CAF1 3800 120 - 1 CCGUUGCUUGUUACACGUCUAAUCUGUUAAAUCCUGUUUACCAAGAUUACAAAUCAAUUGUGGCCAGGAAAUAAAAACACAUGUGGUCCCACACCCCACACCCCCCUCCCAUCCCCCCAC ..(((..((((((((.(......))))....(((((..(((...(((.....)))....)))..)))))))))).)))...(((((.........))))).................... ( -14.90) >DroYak_CAF1 8042 97 - 1 CCGUUGCUUGUUACACGUCUAAUCUGUUAAAUCCUGUUUACCAAGAUUACAAAUCAAUUGUGGCCAGGAAAUAAAAACACAUGUGGUCCCACAC-----------------------CAC ..(((..((((((((.(......))))....(((((..(((...(((.....)))....)))..)))))))))).)))....(((((.....))-----------------------))) ( -14.60) >consensus CCGUUGCUUGUUACACGUCUAAUCUGUUAAAUCCUGUUUACCAAGAUUACAAAUCAAUUGUGGCCAGGAAAUAAAAACACAUGUGGUCCCACAC_______________________CAC ..(((..((((((((.(......))))....(((((..(((...(((.....)))....)))..)))))))))).)))...((((....))))........................... (-14.30 = -14.30 + -0.00)

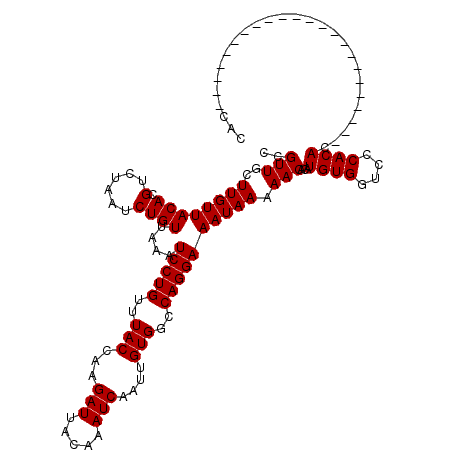

| Location | 20,811,755 – 20,811,875 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -25.83 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20811755 120 + 22224390 GUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCAACGGGCGACCGAUAAUCGCACACACACGCACACCAUCAACAAAA ((((..(((((...(.(((....((((((((((((((((.....)))))))..))))))))).(((((.....)).)))))).)..)))))..))))....................... ( -27.60) >DroEre_CAF1 3840 117 + 1 GUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCAACGGGCGACCGAUAAUCGCAUACACACGCACACCACC---AAAA ((((...................((((((((((((((((.....)))))))..)))))))))..(((((........(((....))).........)))))..))))......---.... ( -26.33) >DroYak_CAF1 8059 117 + 1 GUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCAACGGGCAACCGAUAAUCGCAUACACACGCACACCACC---AAAC ((((...................((((((((((((((((.....)))))))..)))))))))..(((((........(((....))).........)))))..))))......---.... ( -27.33) >consensus GUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCAACGGGCGACCGAUAAUCGCAUACACACGCACACCACC___AAAA ((((...................((((((((((((((((.....)))))))..)))))))))..(((((........(((....))).........)))))..))))............. (-25.83 = -26.16 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:40 2006