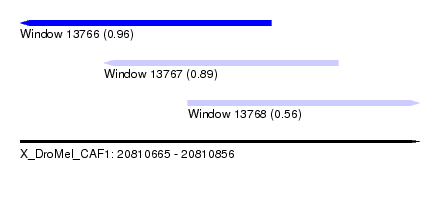
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,810,665 – 20,810,856 |
| Length | 191 |
| Max. P | 0.958428 |

| Location | 20,810,665 – 20,810,785 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -32.38 |
| Energy contribution | -32.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20810665 120 - 22224390 AAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGACGCCAUGUGCACUCCAAUGAUUUUCCGAUCCGCGCGAUUUGCCCGCCAAAUCGCUGGCCAUGUAA ...((((..((((((...(((...((((((((((......))))))))))...)))....))))))......................((((((((.....))))))))))))....... ( -32.20) >DroSim_CAF1 6273 120 - 1 AAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGACGCCAUGUGCACUCCAAUGAUUUUCCGAGCCGCGCGAUUUGCCCGCCAAAUCGCUGGCCAUGUAA ......(((((((((...(((...((((((((((......))))))))))...)))....)))))................(.((((.((((((((.....))))))))))))))))).. ( -34.60) >DroEre_CAF1 2642 120 - 1 AAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGAUGCGAUGUGCACUCCAAUGAUUUUCCGAGCCGCGCGAUUUGCCCGCCAAAUCGCUGGCCAUGUAA ..(((.((((((((....((....((((((((((......)))))))))).)).)))).)))))))...............(.((((.((((((((.....)))))))))))))...... ( -35.50) >DroYak_CAF1 6897 120 - 1 AAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGAUGCGAUGUGCACUCCAAUGAUUUUCCGAGCCGCGCGAUUUGCCCCUCAAAUCGCUGGCCAUGUAA ..(((.((((((((....((....((((((((((......)))))))))).)).)))).)))))))...............(.((((.((((((((.....)))))))))))))...... ( -35.30) >consensus AAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGACGCCAUGUGCACUCCAAUGAUUUUCCGAGCCGCGCGAUUUGCCCGCCAAAUCGCUGGCCAUGUAA ......(((((....(((((((...(((((((((......)))))))))(((((......))))).......)))))))....((((.((((((((.....))))))))))))))))).. (-32.38 = -32.62 + 0.25)


| Location | 20,810,705 – 20,810,817 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20810705 112 - 22224390 GGGCAACAUGCAAAACA--UCGGGAAA------UUGCCAAAAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGACGCCAUGUGCACUCCAAUGAUUUUC .((((....((((....--........------)))).....))))...((((((...(((...((((((((((......))))))))))...)))....)))))).............. ( -24.20) >DroSim_CAF1 6313 112 - 1 GGCCAACAUGCAAAACA--UCGGGAAA------UUGCCAAAAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGACGCCAUGUGCACUCCAAUGAUUUUC (((......((((....--........------))))......)))...((((((...(((...((((((((((......))))))))))...)))....)))))).............. ( -23.00) >DroEre_CAF1 2682 112 - 1 GGCCAACAUGCAAAACA--GCGGGCCA------UGGCCAAAAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGAUGCGAUGUGCACUCCAAUGAUUUUC (((((...(((......--))).....------)))))....(((.((((((((....((....((((((((((......)))))))))).)).)))).))))))).............. ( -30.40) >DroYak_CAF1 6937 120 - 1 GGCCAACAUGCAAAACAUCGCGGGCCAUUGCAAUGGCCAAAAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGAUGCGAUGUGCACUCCAAUGAUUUUC ........(((...((((((((((((((....))))))...((((.....))))..........((((((((((......)))))))))).....))))))))))).............. ( -33.80) >consensus GGCCAACAUGCAAAACA__GCGGGAAA______UGGCCAAAAUGCCACAUGCAUAAAAGUUAACCAAGUUGAUAUUUAAUUGUCAAUUUGCACGACGCCAUGUGCACUCCAAUGAUUUUC ((...((((((..(((......(((((......)))))...((((.....))))....)))...((((((((((......))))))))))......)).)))).....)).......... (-23.38 = -23.38 + 0.00)

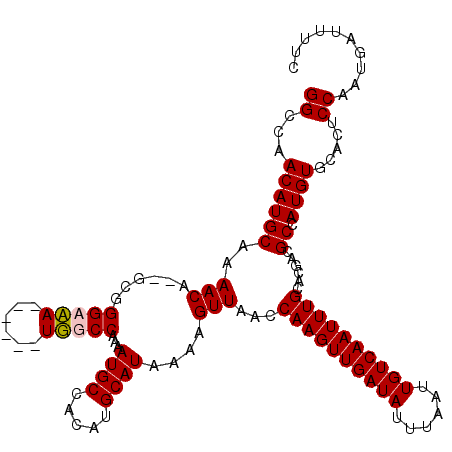
| Location | 20,810,745 – 20,810,856 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20810745 111 + 22224390 AUUAAAUAUCAACUUGGUUAACUUUUAUGCAUGUGGCAUUUUGGCAA------UUUCCCGA--UGUUUUGCAUGUUGCCCAGUGUUGAUUAACAGCUAAUUAAUGG-GGCAGGGCUUUAA ...............((((......((((((...((((((..((...------...)).))--)))).))))))((((((...((((((((.....)))))))).)-))))))))).... ( -24.80) >DroSim_CAF1 6353 111 + 1 AUUAAAUAUCAACUUGGUUAACUUUUAUGCAUGUGGCAUUUUGGCAA------UUUCCCGA--UGUUUUGCAUGUUGGCCAGUGUUGAUUAACAGCUAAUUAAUGG-GGCAGGGCUUUAA .......(((((((((((((((....(((((...((((((..((...------...)).))--)))).)))))))))))))).))))))..............(((-(((...)))))). ( -27.10) >DroEre_CAF1 2722 111 + 1 AUUAAAUAUCAACUUGGUUAACUUUUAUGCAUGUGGCAUUUUGGCCA------UGGCCCGC--UGUUUUGCAUGUUGGCCAGUGUUGAUUAACGGCUAAUUAAUGG-GGCAGGGCUUUAA .......(((((((((((((((....(((((.(..((.....(((..------..))).))--..)..)))))))))))))).))))))..............(((-(((...)))))). ( -30.70) >DroYak_CAF1 6977 120 + 1 AUUAAAUAUCAACUUGGUUAACUUUUAUGCAUGUGGCAUUUUGGCCAUUGCAAUGGCCCGCGAUGUUUUGCAUGUUGGCCAGUGUUGAUUAACGGCUAAUUAAUGGGGGCAGGGCUUUAA .......(((((((((((((((....(((((...((((((..((((((....))))))...)))))).)))))))))))))).)))))).....(((..(....)..))).......... ( -36.10) >consensus AUUAAAUAUCAACUUGGUUAACUUUUAUGCAUGUGGCAUUUUGGCAA______UGGCCCGA__UGUUUUGCAUGUUGGCCAGUGUUGAUUAACAGCUAAUUAAUGG_GGCAGGGCUUUAA .......(((((((((((((((....(((((...((((....(((((......))))).....)))).)))))))))))))).))))))....((((......((....)).)))).... (-27.40 = -27.40 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:36 2006