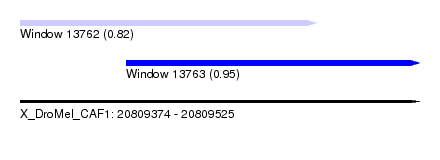
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,809,374 – 20,809,525 |
| Length | 151 |
| Max. P | 0.945209 |

| Location | 20,809,374 – 20,809,486 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20809374 112 + 22224390 AACUAAUCUGUUGAGGUGACGGCUCUUAAAUGGGUUGUGUCAAUAAAUUGGAAAAUAUG-------UGUCAUUGGCCUA-AGGCUUAUUGACUUACGUACACUUCGCAUUGCCCAAAAUA ........(((.(((((((((((((......))))))).................((((-------(((((..((((..-.))))...))))..))))))))))))))............ ( -27.00) >DroSim_CAF1 5057 106 + 1 AACUAAUCUGU-GCAGUGACGGCUCUUAAAUGGGUUGUGUCAAUAAAUUGGAAAAUAUGUGAAUAGUGUCAUUGGCCUA-AGGCCCAUUGACUUACAUA-----------GUCCAAAA-A ...........-....(((((((((......))))..))))).....(((((...(((((((.....((((..((((..-.))))...)))))))))))-----------.)))))..-. ( -28.10) >DroYak_CAF1 5560 110 + 1 UACUAAACUGU-CAAUUGACGGCUCUUAAAUGGCUUGUGGAAGUAUAUUA-AAAAUAUG-------UGUCAUUGGCCAAAAGUCCCAGUGACAUACGUACACUUCAUAAAGGCCAAAA-A .......((((-(....)))))........((((((...(((((.(((..-......((-------(((((((((.........))))))))))).))).)))))....))))))...-. ( -30.80) >consensus AACUAAUCUGU_GAAGUGACGGCUCUUAAAUGGGUUGUGUCAAUAAAUUGGAAAAUAUG_______UGUCAUUGGCCUA_AGGCCCAUUGACUUACGUACACUUC__A__GCCCAAAA_A ...............(..(((((((......)))))))..)..........................((((..((((....))))...))))............................ (-14.50 = -14.50 + 0.00)

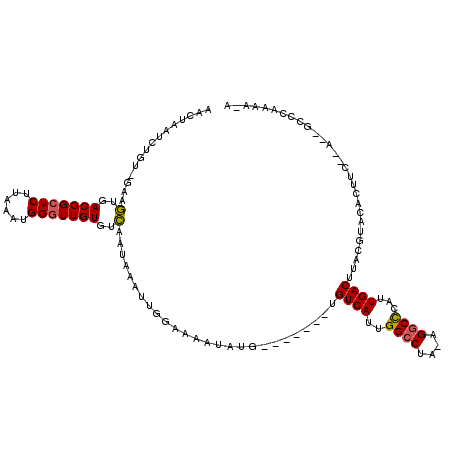
| Location | 20,809,414 – 20,809,525 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.81 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -16.60 |
| Energy contribution | -17.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20809414 111 + 22224390 CAAUAAAUUGGAAAAUAUG-------UGUCAUUGGCCUA-AGGCUUAUUGACUUACGUACACUUCGCAUUGCCCAAAAUAUCAAGUGGCAA-UCCCAGUUUCAUAGGGUAAGCCCCAUAC ....(((((((....((((-------(((((..((((..-.))))...))))..)))))........(((((((..........).)))))-).)))))))....(((....)))..... ( -26.00) >DroSim_CAF1 5096 106 + 1 CAAUAAAUUGGAAAAUAUGUGAAUAGUGUCAUUGGCCUA-AGGCCCAUUGACUUACAUA-----------GUCCAAAA-AUCAAGUGGCAAAGCCCAGUUUCAUAGGGUAAGCCC-AUAC .......(((((...(((((((.....((((..((((..-.))))...)))))))))))-----------.)))))..-.....(.(((...((((.........))))..))))-.... ( -29.90) >DroEre_CAF1 1452 95 + 1 ----------------ACG-------UGUCAUUGGCCAAAAGACCCAAUGACUUGCGUGCACUUCAUAAAGCCCAAAA-AUCAAGUGGCAAAGCCCAGUUUCAUAGGGUAAGCCC-AUAC ----------------(((-------(((((((((.........))))))))..))))((..........((((....-.....).)))...((((.........))))..))..-.... ( -21.80) >DroYak_CAF1 5599 110 + 1 AAGUAUAUUA-AAAAUAUG-------UGUCAUUGGCCAAAAGUCCCAGUGACAUACGUACACUUCAUAAAGGCCAAAA-AUCAAGUGGCAAAGCCCGGUUUCAUAGGGUAAGCCC-AUGC ..((((....-...(..((-------(((((((((.........)))))))))))..)....................-.....(.(((...((((.........))))..))))-)))) ( -24.00) >consensus CAAUAAAUUG_AAAAUAUG_______UGUCAUUGGCCAA_AGGCCCAUUGACUUACGUACACUUCAUAAAGCCCAAAA_AUCAAGUGGCAAAGCCCAGUUUCAUAGGGUAAGCCC_AUAC ...........................((((((((((....))).)))))))..................................(((...((((.........))))..)))...... (-16.60 = -17.60 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:32 2006