
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,797,225 – 20,797,365 |
| Length | 140 |
| Max. P | 0.997349 |

| Location | 20,797,225 – 20,797,338 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -51.10 |
| Consensus MFE | -40.70 |
| Energy contribution | -41.30 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20797225 113 - 22224390 UG------CAGCAGCUGCAGUUGUUGCUACGAUUGUUGCUGCAGCUGUGCGCCUAAUGAGGAGGGGCGGCCAUGCGGCACGUAGCAUCCAUCGGUGGCAACAUCGGGCAAAUAAUCAGC .(------((.((((((((((....((.......)).)))))))))))))((((...(((((...((((((....))).)))....))).))((((....))))))))........... ( -49.00) >DroSec_CAF1 125711 111 - 1 --------UAGCAGCUGCAGUUGUUGCUACGAUUGUUGCUGCAGCUGUGCACCUAAUGAGGAUGGGCGGCCAUGCGGCACGUAGCAUCCAUCGGUGGCAACAUCGGGCAAAUAAUCAGC --------..(((((.((((((((....)))))))).))))).(((((((.((....(((((((.((((((....))).)))..))))).))((((....)))))))))......)))) ( -49.60) >DroSim_CAF1 62754 119 - 1 UGCAGCAGCAGCAGCUGCAGUUGUUGCUACGAUUGUUGCUGCAGCUGUGCGCCUAAUGAGGAGGGGCGGCCAUGCGGCACGUAGCAUCCAUCGGUGGCAACAUCGGGCAAAUAAUCAGC .(((.((((.(((((.((((((((....)))))))).))))).)))))))((((...(((((...((((((....))).)))....))).))((((....))))))))........... ( -54.90) >DroEre_CAF1 48424 119 - 1 UGCAGCAGCAGCAGCUGCAGUUGUUGCUGCGAUUGUAGUUGCAGCUGUGCGCCUAAUGAGGAGGGGCGACCAUGCGGCACGUAGCAUCCAUCGGUGGCAACAUCGGGCAAAUAAUCAGC .(((.((((.((((((((((((((....)))))))))))))).)))))))((((...(((((.((....)).(((........)))))).))((((....))))))))........... ( -53.70) >DroYak_CAF1 100655 110 - 1 UGCA---------GCUGCAGUUGUUGCUGCGAUUGUUGCUGCAGCUGUGCGCCUAAUGAGUAGGGGCGGCCAUGCGGCACGUAGCAUCCAUCGGUGGCAACAUCGGGCAAAUAAUCAGC ((((---------((.((((((((....)))))))).))))))(((((((.((((.....)))).)))(((((((........))))....(((((....)))))))).......)))) ( -48.30) >consensus UGCA____CAGCAGCUGCAGUUGUUGCUACGAUUGUUGCUGCAGCUGUGCGCCUAAUGAGGAGGGGCGGCCAUGCGGCACGUAGCAUCCAUCGGUGGCAACAUCGGGCAAAUAAUCAGC ..........(((((.((((((((....)))))))).))))).((((..(((((.........)))))(((((((........))))....(((((....)))))))).......)))) (-40.70 = -41.30 + 0.60)



| Location | 20,797,264 – 20,797,365 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -25.07 |
| Energy contribution | -26.11 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20797264 101 + 22224390 GUGCCGCAUGGCCGCCCCUCCUCAUUAGGCGCACAGCUGCAGCAACAAUCGUAGCAACAACUGCAGCUGCUG------CAACUCCAUGUGCAACAUACAAAACGCAG--------- .(((.((((((..(((...........)))((((((((((((..................))))))))).))------)....)))))))))...............--------- ( -32.97) >DroSec_CAF1 125750 95 + 1 GUGCCGCAUGGCCGCCCAUCCUCAUUAGGUGCACAGCUGCAGCAACAAUCGUAGCAACAACUGCAGCUGCUA------------CAUCUGCAACAUGCAAAGCGCAG--------- ((((.(((((...((...........(((((..(((((((((..................)))))))))...------------)))))))..)))))...))))..--------- ( -29.87) >DroSim_CAF1 62793 107 + 1 GUGCCGCAUGGCCGCCCCUCCUCAUUAGGCGCACAGCUGCAGCAACAAUCGUAGCAACAACUGCAGCUGCUGCUGCUGCAACUCCAUCUGCAACAUGCAAAGCGCAG--------- ((((.((((((((..............)))(((((((.(((((.((....)).(((.....))).))))).)))).)))..............)))))...))))..--------- ( -38.24) >DroEre_CAF1 48463 116 + 1 GUGCCGCAUGGUCGCCCCUCCUCAUUAGGCGCACAGCUGCAACUACAAUCGCAGCAACAACUGCAGCUGCUGCUGCUGCAACUCCAUCUGCAACAUGCAAAGCGCUGCACCUACUG (((((....((........))......)))))...(((((..........)))))......((((((.(((((((.((((........)))).)).))..)))))))))....... ( -36.90) >DroYak_CAF1 100694 107 + 1 GUGCCGCAUGGCCGCCCCUACUCAUUAGGCGCACAGCUGCAGCAACAAUCGCAGCAACAACUGCAGC---------UGCAACUCCAUUUGCAACAUGCAGAGGGCAGCAUCUGCUG ..((.((.((..((((...........))))..)))).))((((......((((......)))).((---------(((....((.(((((.....))))))))))))...)))). ( -35.50) >consensus GUGCCGCAUGGCCGCCCCUCCUCAUUAGGCGCACAGCUGCAGCAACAAUCGUAGCAACAACUGCAGCUGCUG____UGCAACUCCAUCUGCAACAUGCAAAGCGCAG_________ ((((.((((((.((((...........))))).(((((((((..................)))))))))........................)))))...))))........... (-25.07 = -26.11 + 1.04)

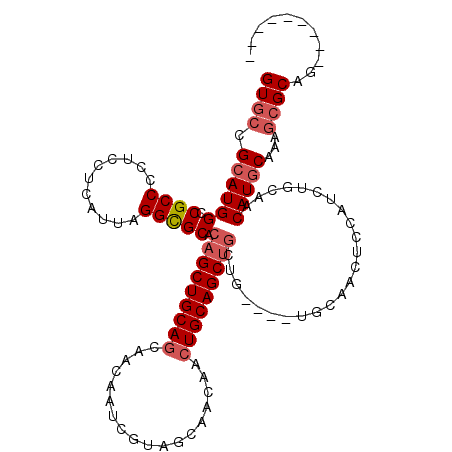

| Location | 20,797,264 – 20,797,365 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -46.28 |
| Consensus MFE | -32.10 |
| Energy contribution | -32.78 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20797264 101 - 22224390 ---------CUGCGUUUUGUAUGUUGCACAUGGAGUUG------CAGCAGCUGCAGUUGUUGCUACGAUUGUUGCUGCAGCUGUGCGCCUAAUGAGGAGGGGCGGCCAUGCGGCAC ---------............(((((((..((((((((------(((((...((((((((....)))))))))))))))))).(((.(((.......))).))).)))))))))). ( -41.20) >DroSec_CAF1 125750 95 - 1 ---------CUGCGCUUUGCAUGUUGCAGAUG------------UAGCAGCUGCAGUUGUUGCUACGAUUGUUGCUGCAGCUGUGCACCUAAUGAGGAUGGGCGGCCAUGCGGCAC ---------.(((.....)))(((((((....------------..(((((.((((((((....)))))))).))))).(((((.(((((....))).)).)))))..))))))). ( -40.30) >DroSim_CAF1 62793 107 - 1 ---------CUGCGCUUUGCAUGUUGCAGAUGGAGUUGCAGCAGCAGCAGCUGCAGUUGUUGCUACGAUUGUUGCUGCAGCUGUGCGCCUAAUGAGGAGGGGCGGCCAUGCGGCAC ---------(((((...(((.((((((((......)))))))))))(((((.((((((((....)))))))).))))).(((((.(.(((....)))..).)))))..)))))... ( -46.10) >DroEre_CAF1 48463 116 - 1 CAGUAGGUGCAGCGCUUUGCAUGUUGCAGAUGGAGUUGCAGCAGCAGCAGCUGCAGUUGUUGCUGCGAUUGUAGUUGCAGCUGUGCGCCUAAUGAGGAGGGGCGACCAUGCGGCAC .....(.((((.(((((((((((((((((.((.(((((((((((((((.......))))))))))))))).)).))))))).)))).(((....))).))))))....)))).).. ( -53.40) >DroYak_CAF1 100694 107 - 1 CAGCAGAUGCUGCCCUCUGCAUGUUGCAAAUGGAGUUGCA---------GCUGCAGUUGUUGCUGCGAUUGUUGCUGCAGCUGUGCGCCUAAUGAGUAGGGGCGGCCAUGCGGCAC ..(((((........))))).(((((((..((((((((((---------((.((((((((....)))))))).))))))))).(((.((((.....)))).))).)))))))))). ( -50.40) >consensus _________CUGCGCUUUGCAUGUUGCAGAUGGAGUUGCA____CAGCAGCUGCAGUUGUUGCUACGAUUGUUGCUGCAGCUGUGCGCCUAAUGAGGAGGGGCGGCCAUGCGGCAC ...........((.....)).(((((((..(((.............(((((((((((....((.......)).))))))))))).(((((.........))))).)))))))))). (-32.10 = -32.78 + 0.68)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:25 2006