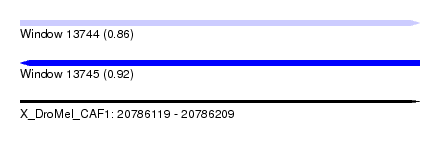
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,786,119 – 20,786,209 |
| Length | 90 |
| Max. P | 0.919097 |

| Location | 20,786,119 – 20,786,209 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 88.51 |
| Mean single sequence MFE | -13.07 |
| Consensus MFE | -9.25 |
| Energy contribution | -8.88 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
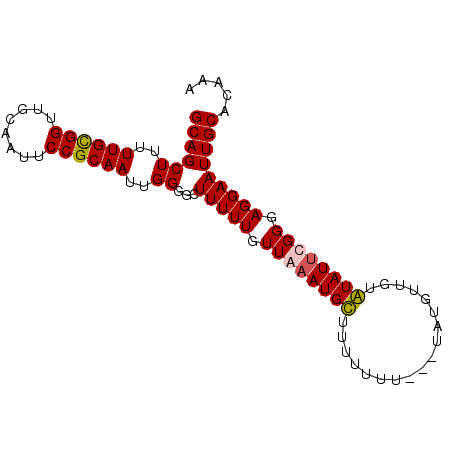
>X_DroMel_CAF1 20786119 90 + 22224390 UUUGUGCAAUUCCUCCCGUAUAUACAACAUACAAAAAAAAAACAUUUAACAAAAACCCCCAAUUGCGGAAUUGCAACCACAAAAAGCUGC ....(((((((((....((((.......)))).................(((..........))).)))))))))............... ( -12.30) >DroSec_CAF1 114778 86 + 1 UUUGCGCAAUUCCUCCCAAAUACACAACAUA---AAA-AAAGCAUUUAACAAAAAGCCCCAAUUGCGGAAUUGAAACCGCAAAAAGCUGC (((((((((((((................((---((.-......)))).......((.......)))))))))....))))))....... ( -12.30) >DroSim_CAF1 50611 87 + 1 UUUGUGCAAUUCCUCCCGAAUACACAACAUA---AAAAAAAGCAUUUAACAAAAAGCCCCAAUUGCGGAAUUGAAACCGCAAAAAGCUGC .(((((..((((.....)))).)))))....---....................(((.....((((((........))))))...))).. ( -13.90) >DroEre_CAF1 37433 82 + 1 UUUGUGCAAUUCCUCCCAAAUAUACAAAA--------AAAAACAUUUAACAAAAAGCACCAAUUGCGGAAUUGCAACCGCAAAAAGCUGC ....(((((((((................--------.......(((....))).(((.....))))))))))))...(((......))) ( -13.80) >consensus UUUGUGCAAUUCCUCCCAAAUACACAACAUA___AAAAAAAACAUUUAACAAAAAGCCCCAAUUGCGGAAUUGAAACCGCAAAAAGCUGC (((((((((((((.....................................................)))))))....))))))....... ( -9.25 = -8.88 + -0.38)


| Location | 20,786,119 – 20,786,209 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 88.51 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20786119 90 - 22224390 GCAGCUUUUUGUGGUUGCAAUUCCGCAAUUGGGGGUUUUUGUUAAAUGUUUUUUUUUUGUAUGUUGUAUAUACGGGAGGAAUUGCACAAA ((((((......))))))((((((.......)))))).........(((.((((((((((((((...))))))))))))))..))).... ( -21.00) >DroSec_CAF1 114778 86 - 1 GCAGCUUUUUGCGGUUUCAAUUCCGCAAUUGGGGCUUUUUGUUAAAUGCUUU-UUU---UAUGUUGUGUAUUUGGGAGGAAUUGCGCAAA ((((((((((((((........))))))..)))))(((((.((((((((...-...---........)))))))).))))).)))..... ( -22.14) >DroSim_CAF1 50611 87 - 1 GCAGCUUUUUGCGGUUUCAAUUCCGCAAUUGGGGCUUUUUGUUAAAUGCUUUUUUU---UAUGUUGUGUAUUCGGGAGGAAUUGCACAAA (((.((..((((((........))))))..))(((.....)))...))).......---....((((((((((....)))..))))))). ( -21.90) >DroEre_CAF1 37433 82 - 1 GCAGCUUUUUGCGGUUGCAAUUCCGCAAUUGGUGCUUUUUGUUAAAUGUUUUU--------UUUUGUAUAUUUGGGAGGAAUUGCACAAA ((((((..((((((........))))))..)))..(((((.((((((((....--------......)))))))).))))).)))..... ( -20.40) >consensus GCAGCUUUUUGCGGUUGCAAUUCCGCAAUUGGGGCUUUUUGUUAAAUGCUUUUUUU___UAUGUUGUAUAUUCGGGAGGAAUUGCACAAA ((((((..((((((........))))))..))...(((((.((((((((..................)))))))).)))))))))..... (-17.96 = -18.02 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:15 2006