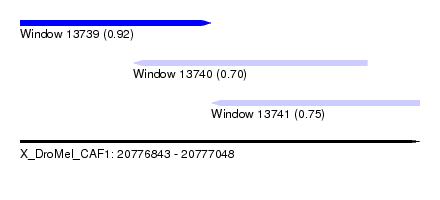
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,776,843 – 20,777,048 |
| Length | 205 |
| Max. P | 0.915840 |

| Location | 20,776,843 – 20,776,941 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
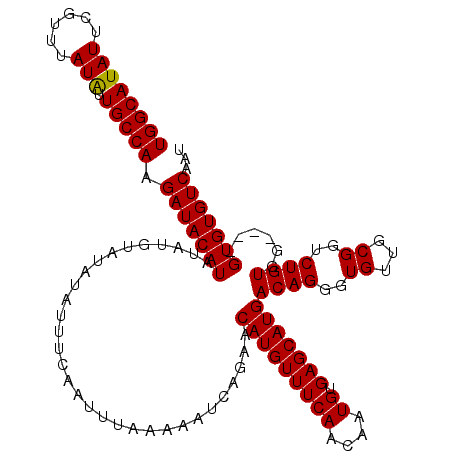
>X_DroMel_CAF1 20776843 98 + 22224390 UGGCAUAUUCGUUUAUAUUGCCAAGAUACAUA----------------------AAUUAGAACAUGUUUCAAUAAUGUGAGCAUGACAGGGUGUUGCGGUCUGUGCGUGCGUGUGUCAAU ((((((((......))).))))).(((((((.----------------------........((((((.....)))))).((((((((((.((...)).))))).))))))))))))... ( -29.70) >DroSec_CAF1 105479 116 + 1 UGGCAUAUUCGUUUAUAUUGCCAAGAUACAUAUAUGUAUAUAUUUCAAUUUAAAAAUCAGAACAUGUUUCAACAAUGUGAGCAUGACAGGGUGUUGCGUUCUGUGG----GUGUGUCAAU ((((((((((((((((((((.....(((((....)))))....................(((.....)))..))))))))))...((((..((...))..))))))----)))))))).. ( -29.20) >DroSim_CAF1 40955 116 + 1 UGGCAUAUUCGUUUAUGUUGCCAAGAUACAUAUAUGUAUAUAUUUCAAUUUAAAAAUCAGAACAUGUUUCAACAAUGUGAGCAUGACAGGGUGUUGCGGUCUGUGG----GUGUGUCAAU ((((((((((((((((((((.....(((((....)))))....................(((.....)))..))))))))))...(((((.((...)).)))))))----)))))))).. ( -27.00) >consensus UGGCAUAUUCGUUUAUAUUGCCAAGAUACAUAUAUGUAUAUAUUUCAAUUUAAAAAUCAGAACAUGUUUCAACAAUGUGAGCAUGACAGGGUGUUGCGGUCUGUGG____GUGUGUCAAU ((((((((......))).))))).(((((((...............................(((((((((....)).)))))))((((..((...))..))))......)))))))... (-24.35 = -24.13 + -0.22)

| Location | 20,776,901 – 20,777,021 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.76 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20776901 120 - 22224390 CGUACAGCCACACGGCAAUAAAAAAUAUAGAAAAUUGGAAUUCAACAUGACGCAUACGCAGUGAAAGUAUGUUUCAAUUCAUUGACACACGCACGCACAGACCGCAACACCCUGUCAUGC ......(((....))).............................((((((((....)).(((...(..(((.((((....)))).)))..)...)))...............)))))). ( -23.00) >DroSec_CAF1 105559 116 - 1 CGUACAGUCGCAAGAAUAUACAAAAUAUAAAUAACUGGAAUUAAGCAUGACGCAUACGCAGUGAAAGUAUGUUUCAAUUCAUUGACACAC----CCACAGAACGCAACACCCUGUCAUGC .(((...((....))...))).......................(((((((((....)).......(..(((.((((....)))).))).----.).................))))))) ( -21.10) >DroSim_CAF1 41035 116 - 1 CGUACAGUCGCAAGACUAUACAAAAUACAGAAAACUGGAAUUAAGCAUGACGCAUACGCAGUGAAAGUGUGUUUCAAUUCAUUGACACAC----CCACAGACCGCAACACCCUGUCAUGC .(((.((((....)))).)))......(((....))).......(((((((((....)).(((...(((((((..........)))))))----.)))...............))))))) ( -33.40) >consensus CGUACAGUCGCAAGACUAUACAAAAUAUAGAAAACUGGAAUUAAGCAUGACGCAUACGCAGUGAAAGUAUGUUUCAAUUCAUUGACACAC____CCACAGACCGCAACACCCUGUCAUGC .(((.((((....)))).))).......................(((((((((....)).(((......(((.((((....)))).)))......)))...............))))))) (-21.38 = -22.27 + 0.89)


| Location | 20,776,941 – 20,777,048 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.16 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.00 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20776941 107 - 22224390 UUGAGUUAGUCAUUCCCAUUAGUAUUUCGUACAGCCACACGGCAAUAAAAAAUAUAGAAAAUUGGAAUUCAACAUGACGCAUACGCAGUGAAAGUAUGUUUCAAUUC ....(((.(..(((((.(((.((((((..((..(((....)))..))..))))))....))).))))).)))).(((.((((((.........)))))).))).... ( -19.70) >DroSec_CAF1 105595 107 - 1 UAGAGUUAUUCACCACCAUUAGUAUUUCGUACAGUCGCAAGAAUAUACAAAAUAUAAAUAACUGGAAUUAAGCAUGACGCAUACGCAGUGAAAGUAUGUUUCAAUUC ..(((((.....(((..(((.((((((.(((...((....))...))).)))))).)))...)))..........((.((((((.........)))))).))))))) ( -20.10) >DroSim_CAF1 41071 107 - 1 UAGAGUUAUUCACCACCAUUAGUAUUUCGUACAGUCGCAAGACUAUACAAAAUACAGAAAACUGGAAUUAAGCAUGACGCAUACGCAGUGAAAGUGUGUUUCAAUUC ..(((((.....(((...((.((((((.(((.((((....)))).))).)))))).))....)))..........((.((((((.........)))))).))))))) ( -25.30) >consensus UAGAGUUAUUCACCACCAUUAGUAUUUCGUACAGUCGCAAGACUAUACAAAAUAUAGAAAACUGGAAUUAAGCAUGACGCAUACGCAGUGAAAGUAUGUUUCAAUUC ..(((((........(((((.((((((.(((.((((....)))).))).)))))).))....)))..........((.((((((.........)))))).))))))) (-20.10 = -20.00 + -0.10)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:12 2006