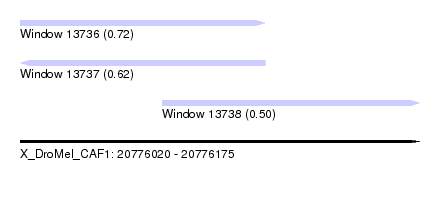
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,776,020 – 20,776,175 |
| Length | 155 |
| Max. P | 0.721704 |

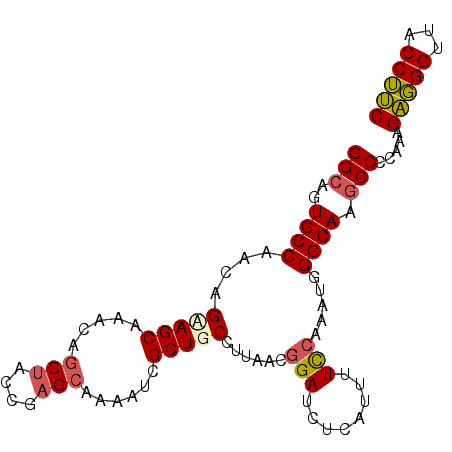
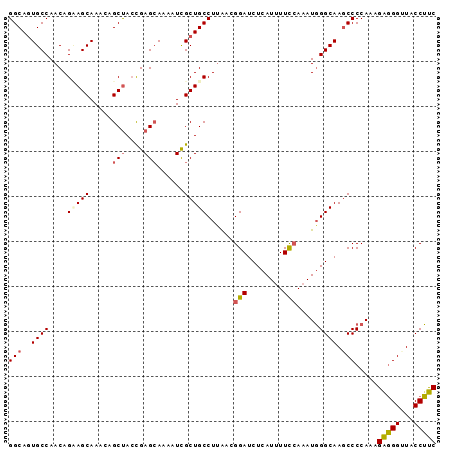
| Location | 20,776,020 – 20,776,115 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 89.68 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -20.72 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
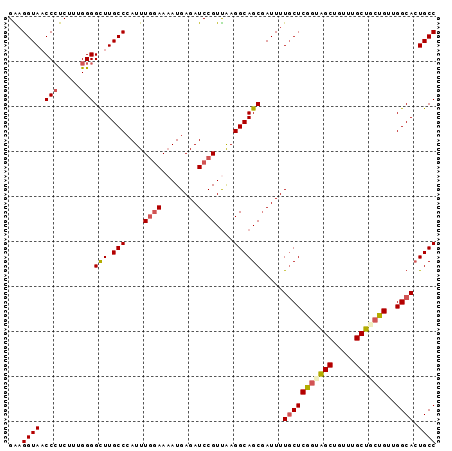
>X_DroMel_CAF1 20776020 95 + 22224390 GGCACUGCCAACAGCAGCAAAAAGCUACCGAGCAAAAUCGCUGCCUUAACGGAUCUCAUAUUCCAAAUGGGCAAGCCCCAAAGAGGGGUACCUUC ((...((((....(((((.....(((....)))......)))))......(((........))).....)))).(((((.....))))).))... ( -28.50) >DroSec_CAF1 89908 95 + 1 GGCAGUGCCAACAGAAGCAAACAGCUACCGAGCAAAAUCGCUGCCUUAACGGAUCUCAUUUUCCAAAUGGGCAAGCCCCAAAGAGGGGUACCUUC ((((((((........)).....(((....)))......)))))).....((..((((((.....))))))...(((((.....))))).))... ( -26.90) >DroSim_CAF1 35226 95 + 1 GGCAGUGCCAACAGAAGCAAACAGCUACUGAGCAAAAUCGCUGCCUUAACGGAUCUCAUUUUCCAAAUGGGCAAGCCCCAAAGAGGGUUACCUUC (((((((....(((.(((.....))).)))........))))))).....(((........)))...((((.....))))..((((....)))). ( -26.80) >DroEre_CAF1 27384 95 + 1 GGCAGUGCCAACAGCAGCAAACUGCCAGCGAGCAAAAUUGCUGCCUUGACGGAUCUCAUUUUCCAAAUUGGCAAGCCCCAAAGGCGGUUACCGCC (((..((((((..(((((((..(((......)))...)))))))......(((........)))...)))))).))).....(((((...))))) ( -35.10) >DroYak_CAF1 77082 95 + 1 GGCAGUGCCAACAGUAGCAAACAGCUACCGAGGAAAAUCGCUGCCUCAACGGAUCUCAUUUUUGAAAUGGGCAAACCCCAACGGGGGUUACCUCC (((((((((....(((((.....)))))...)).....))))))).....((..((((((.....))))))..((((((....)))))).))... ( -31.50) >consensus GGCAGUGCCAACAGAAGCAAACAGCUACCGAGCAAAAUCGCUGCCUUAACGGAUCUCAUUUUCCAAAUGGGCAAGCCCCAAAGAGGGUUACCUUC (((..((((....(((((.....(((....)))......)))))......(((........))).....)))).))).....(((((...))))) (-20.72 = -20.88 + 0.16)

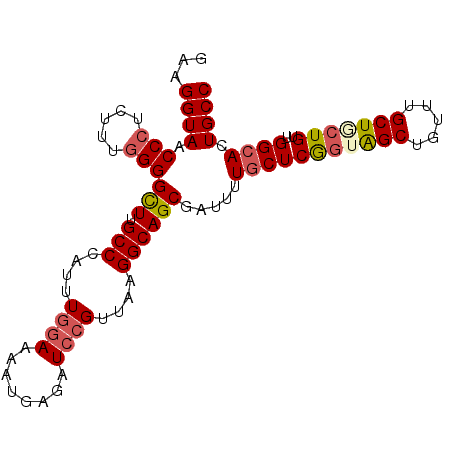
| Location | 20,776,020 – 20,776,115 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 89.68 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -24.58 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20776020 95 - 22224390 GAAGGUACCCCUCUUUGGGGCUUGCCCAUUUGGAAUAUGAGAUCCGUUAAGGCAGCGAUUUUGCUCGGUAGCUUUUUGCUGCUGUUGGCAGUGCC ...((((((((.....)))(((.(((....((((........))))....)))))).....(((((((((((.....)))))))..))))))))) ( -35.20) >DroSec_CAF1 89908 95 - 1 GAAGGUACCCCUCUUUGGGGCUUGCCCAUUUGGAAAAUGAGAUCCGUUAAGGCAGCGAUUUUGCUCGGUAGCUGUUUGCUUCUGUUGGCACUGCC ...((((((((.....))))..)))).....(((........))).....(((((......(((((((.(((.....))).)))..))))))))) ( -27.70) >DroSim_CAF1 35226 95 - 1 GAAGGUAACCCUCUUUGGGGCUUGCCCAUUUGGAAAAUGAGAUCCGUUAAGGCAGCGAUUUUGCUCAGUAGCUGUUUGCUUCUGUUGGCACUGCC ...((((.(((.....)))(((.(((....((((........))))....)))))).....(((((((.(((.....))).)))..)))).)))) ( -28.40) >DroEre_CAF1 27384 95 - 1 GGCGGUAACCGCCUUUGGGGCUUGCCAAUUUGGAAAAUGAGAUCCGUCAAGGCAGCAAUUUUGCUCGCUGGCAGUUUGCUGCUGUUGGCACUGCC (((((...))))).....(((.(((((((..(((........))).....((((((((..(((((....))))).)))))))))))))))..))) ( -39.50) >DroYak_CAF1 77082 95 - 1 GGAGGUAACCCCCGUUGGGGUUUGCCCAUUUCAAAAAUGAGAUCCGUUGAGGCAGCGAUUUUCCUCGGUAGCUGUUUGCUACUGUUGGCACUGCC ((((((((((((....)))).)))))..(((((....)))))))).....(((((.(.....((.(((((((.....)))))))..))).))))) ( -35.60) >consensus GAAGGUAACCCUCUUUGGGGCUUGCCCAUUUGGAAAAUGAGAUCCGUUAAGGCAGCGAUUUUGCUCGGUAGCUGUUUGCUGCUGUUGGCACUGCC ...((((.(((.....)))(((.(((....((((........))))....)))))).....(((((((((((.....)))))))..)))).)))) (-24.58 = -25.42 + 0.84)

| Location | 20,776,075 – 20,776,175 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -20.08 |
| Consensus MFE | -9.61 |
| Energy contribution | -8.62 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
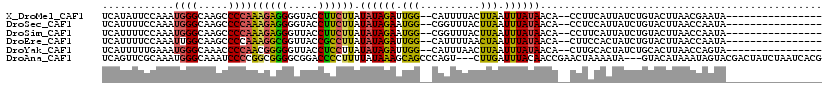
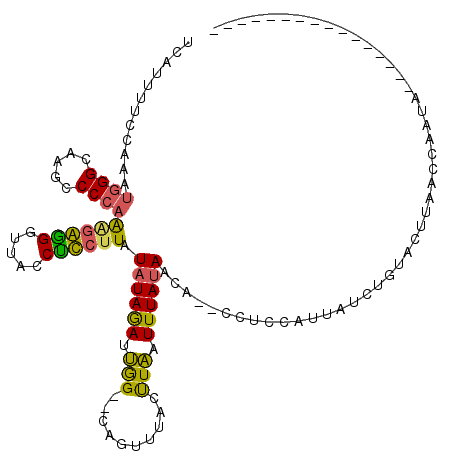
>X_DroMel_CAF1 20776075 100 + 22224390 UCAUAUUCCAAAUGGGCAAGCCCCAAAGAGGGGUACCUUCUUAUAUAGAUUGG--CAUUUUACUUAAUUUAUAACA--CCUUCAUUAUCUGUACUUAACGAAUA---------------- ...(((((.....(..((.(((((.....))))).........((((((((((--........))))))))))...--...........))..).....)))))---------------- ( -16.60) >DroSec_CAF1 89963 100 + 1 UCAUUUUCCAAAUGGGCAAGCCCCAAAGAGGGGUACCUUCUUAUAUAGAAUGG--CGGUUUACUUAAUUUAUAACA--CCUCCAUUAUCUGUACUUAACCAAUA---------------- ............(((....(((((.....))))).........((((((((((--.(((................)--)).))))..)))))).....)))...---------------- ( -21.79) >DroSim_CAF1 35281 100 + 1 UCAUUUUCCAAAUGGGCAAGCCCCAAAGAGGGUUACCUUCUUAUAUAGAAUGG--CGGUUUACUUAAUUUAUAACA--CCUUCAUUAUCUGUACUUAACCAAUA---------------- ............(((..(((.....(((((((...))))))).((((((((((--.(((................)--)).))))..)))))))))..)))...---------------- ( -18.29) >DroEre_CAF1 27439 100 + 1 UCAUUUUCCAAAUUGGCAAGCCCCAAAGGCGGUUACCGCCUUAUAUAGAUUGG--CAUUUUAACUAAUUUAUAACA--CUUCCACUAUCUGUACUUAACCAAUA---------------- ...........(((((.........(((((((...))))))).((((((((((--........))))))))))...--....................))))).---------------- ( -21.20) >DroYak_CAF1 77137 100 + 1 UCAUUUUUGAAAUGGGCAAACCCCAACGGGGGUUACCUCCUUAUAUAGAUUGG--CAUUUAACUUAAUUUAUAACA--CUUGCACUAUCUGCACUUAACCAGUA---------------- .......((..(((((((((((((....)))))).........((((((((((--........))))))))))...--..))).))))...))...........---------------- ( -19.10) >DroAna_CAF1 32963 114 + 1 UCAGUUCGCAAAUGGGCAAAUCCCCGGCGGGGCGGACCCCUUUUAUAAAGCAGCCCAGU---CUUGAUUUACAACCGAACUAAAAUA---GUACAUAAAUAGUACGACUAUCUAAUCACG ..((((((.....(((......)))(((.((((...................)))).))---)............))))))......---((((.......))))............... ( -23.51) >consensus UCAUUUUCCAAAUGGGCAAGCCCCAAAGAGGGUUACCUCCUUAUAUAGAUUGG__CAGUUUACUUAAUUUAUAACA__CCUCCAUUAUCUGUACUUAACCAAUA________________ ............((((.....))))((((((.....)))))).((((((.(((..........))).))))))............................................... ( -9.61 = -8.62 + -1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:09 2006