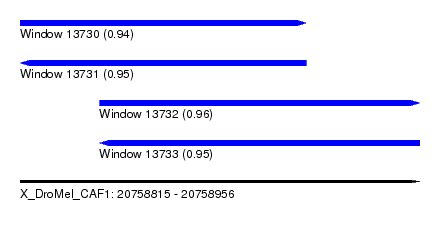
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,758,815 – 20,758,956 |
| Length | 141 |
| Max. P | 0.959979 |

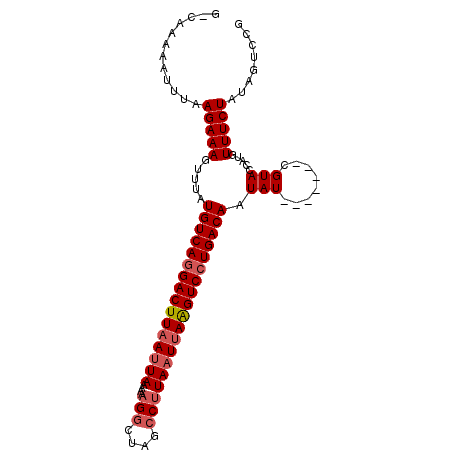
| Location | 20,758,815 – 20,758,916 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.44 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -19.41 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20758815 101 + 22224390 CGGACUUUAGAAACAUGGUACUACAAUAUAUAUUGUCAGGACUUAAUUAGGGCUAGCCUUUUUUAGUUAAGUCGUGACAUAAACUUUCUUAAAUUUUUCCC .(((.((((((((..((......))........(((((.(((((((((((((....)))....)))))))))).))))).....)))).))))....))). ( -23.70) >DroSec_CAF1 78141 93 + 1 CGGACUAUAGAAACAUGGUACG-------AUAUUGUCAGGACCUUAUUAAGUCUAGUCUUUUUUAAUUAAGUCCUGACAUAAACUUUCUUAAAUUUUUG-C ((.(((((......))))).))-------....(((((((((...((((((..........))))))...)))))))))....................-. ( -20.20) >DroSim_CAF1 23486 93 + 1 CGGAGUAUAGAAACAUGGUACG-------AUAUUGUCAGGACUUAAUUAAGGCUAGCCUUUUUUAAUUAAGUCCUGACAUAAGCUUUCUUAAAUUUUUG-C .(((((((.........)))).-------....(((((((((((((((((((....)))....)))))))))))))))).......)))..........-. ( -26.50) >consensus CGGACUAUAGAAACAUGGUACG_______AUAUUGUCAGGACUUAAUUAAGGCUAGCCUUUUUUAAUUAAGUCCUGACAUAAACUUUCUUAAAUUUUUG_C .................................(((((((((((((((((((........)))))))))))))))))))...................... (-19.41 = -20.30 + 0.89)



| Location | 20,758,815 – 20,758,916 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 86.44 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.87 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20758815 101 - 22224390 GGGAAAAAUUUAAGAAAGUUUAUGUCACGACUUAACUAAAAAAGGCUAGCCCUAAUUAAGUCCUGACAAUAUAUAUUGUAGUACCAUGUUUCUAAAGUCCG .(((....((((.((((.....(((((.(((((((.((.....((....)).)).))))))).)))))(((.....))).........)))))))).))). ( -17.90) >DroSec_CAF1 78141 93 - 1 G-CAAAAAUUUAAGAAAGUUUAUGUCAGGACUUAAUUAAAAAAGACUAGACUUAAUAAGGUCCUGACAAUAU-------CGUACCAUGUUUCUAUAGUCCG (-((.((((((....)))))).(((((((((((.(((((............))))).)))))))))))....-------.......)))............ ( -18.70) >DroSim_CAF1 23486 93 - 1 G-CAAAAAUUUAAGAAAGCUUAUGUCAGGACUUAAUUAAAAAAGGCUAGCCUUAAUUAAGUCCUGACAAUAU-------CGUACCAUGUUUCUAUACUCCG (-((......((((....))))((((((((((((((((....(((....)))))))))))))))))))....-------.......)))............ ( -26.00) >consensus G_CAAAAAUUUAAGAAAGUUUAUGUCAGGACUUAAUUAAAAAAGGCUAGCCUUAAUUAAGUCCUGACAAUAU_______CGUACCAUGUUUCUAUAGUCCG ............(((((.....((((((((((((((((....(((....))))))))))))))))))).(((........))).....)))))........ (-18.42 = -19.87 + 1.45)


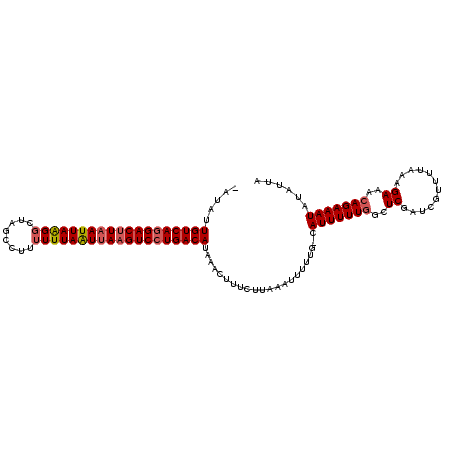
| Location | 20,758,843 – 20,758,956 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.34 |
| Energy contribution | -23.57 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20758843 113 + 22224390 UAUAUUGUCAGGACUUAAUUAGGGCUAGCCUUUUUUAGUUAAGUCGUGACAUAAACUUUCUUAAAUUUUUCCCAUUUUUUGAUCGAUCGUUUUUAGGAAACAGAAAUAUAUUA ((((((((((.(((((((((((((....)))....)))))))))).))))......(((((((((....((.((.....))...)).....)))))))))....))))))... ( -26.30) >DroSec_CAF1 78163 111 + 1 -AUAUUGUCAGGACCUUAUUAAGUCUAGUCUUUUUUAAUUAAGUCCUGACAUAAACUUUCUUAAAUUUUUG-CAUUUUUGGCUCGAUCGUUUUAUAGAAACAGAAAUAUAUUA -....(((((((((...((((((..........))))))...)))))))))....................-.(((((((..((............))..)))))))...... ( -21.80) >DroSim_CAF1 23508 110 + 1 -AUAUUGUCAGGACUUAAUUAAGGCUAGCCUUUUUUAAUUAAGUCCUGACAUAAGCUUUCUUAAAUUUUUG-CAUUUUUGGCUCGAUCGUU-UAAAGAAACAGAAAUAUUUUA -....(((((((((((((((((((....)))....))))))))))))))))...((..............)-)(((((((..((.......-....))..)))))))...... ( -31.04) >consensus _AUAUUGUCAGGACUUAAUUAAGGCUAGCCUUUUUUAAUUAAGUCCUGACAUAAACUUUCUUAAAUUUUUG_CAUUUUUGGCUCGAUCGUUUUAAAGAAACAGAAAUAUAUUA .....(((((((((((((((((((........)))))))))))))))))))......................(((((((..((............))..)))))))...... (-22.34 = -23.57 + 1.23)


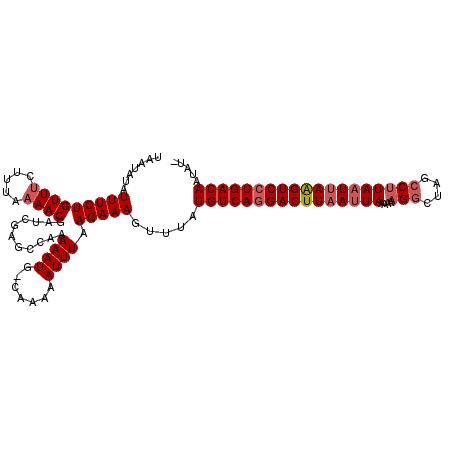
| Location | 20,758,843 – 20,758,956 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -19.45 |
| Energy contribution | -21.23 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20758843 113 - 22224390 UAAUAUAUUUCUGUUUCCUAAAAACGAUCGAUCAAAAAAUGGGAAAAAUUUAAGAAAGUUUAUGUCACGACUUAACUAAAAAAGGCUAGCCCUAAUUAAGUCCUGACAAUAUA .......(((((.(((((((.....((....))......)))))))......))))).....(((((.(((((((.((.....((....)).)).))))))).)))))..... ( -20.00) >DroSec_CAF1 78163 111 - 1 UAAUAUAUUUCUGUUUCUAUAAAACGAUCGAGCCAAAAAUG-CAAAAAUUUAAGAAAGUUUAUGUCAGGACUUAAUUAAAAAAGACUAGACUUAAUAAGGUCCUGACAAUAU- .......(((((((((.....))))......((.......)-).........))))).....(((((((((((.(((((............))))).)))))))))))....- ( -21.70) >DroSim_CAF1 23508 110 - 1 UAAAAUAUUUCUGUUUCUUUA-AACGAUCGAGCCAAAAAUG-CAAAAAUUUAAGAAAGCUUAUGUCAGGACUUAAUUAAAAAAGGCUAGCCUUAAUUAAGUCCUGACAAUAU- .......(((((((((....)-)))......((.......)-).........))))).....((((((((((((((((....(((....)))))))))))))))))))....- ( -28.60) >consensus UAAUAUAUUUCUGUUUCUUUAAAACGAUCGAGCCAAAAAUG_CAAAAAUUUAAGAAAGUUUAUGUCAGGACUUAAUUAAAAAAGGCUAGCCUUAAUUAAGUCCUGACAAUAU_ .......(((((((((.....))))...........((((.......)))).))))).....((((((((((((((((....(((....)))))))))))))))))))..... (-19.45 = -21.23 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:05 2006