| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,329,782 – 2,329,889 |
| Length | 107 |
| Max. P | 0.878921 |

| Location | 2,329,782 – 2,329,889 |
|---|---|
| Length | 107 |
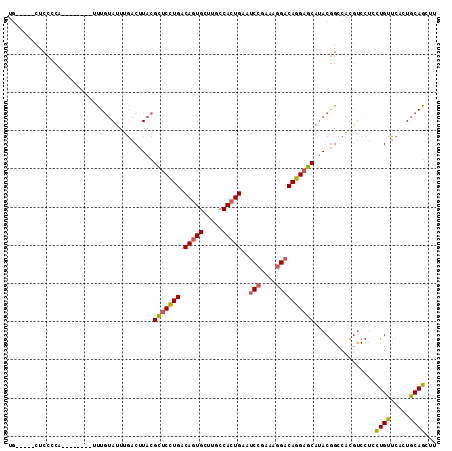
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -29.90 |
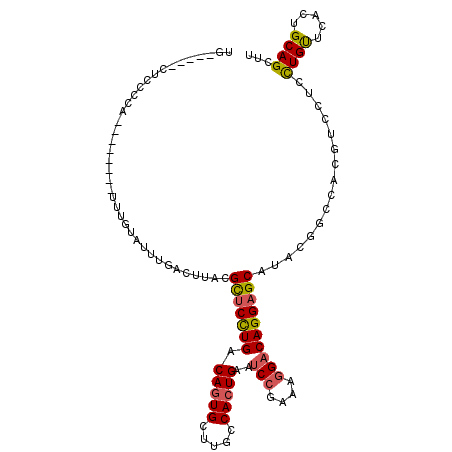
| Consensus MFE | -22.70 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2329782 107 + 22224390 UG-----CUCCCCUAUUUGUAUUUUGUAUUUGACUUACGCUCCUGACAGUGCUUGCCACUGAAUCCGAAAGGACAGGAGCAUACGGCCACGUCCUCCUGUUCACUGCAGCUU ..-----.........((((((...(((.......)))(((((((.(((((.....)))))..(((....))))))))))))))))..........((((.....))))... ( -30.20) >DroSec_CAF1 1036 99 + 1 UG-----CUCCCCU--------UUUGUAUUUGACUUACGUUCCUGACAGUGCUUGCCACUGAAUCCGAAAGGACAGGAGCAUACGGCCACGUCCUCCUGCUCACUGCAGCUU .(-----((.....--------...(((.......)))(((((((.(((((.....)))))..(((....))))))))))....))).........((((.....))))... ( -29.50) >DroSim_CAF1 942 99 + 1 UG-----CUCCCCU--------UUUGUAUUUGACUUACGUUCCUGACAGUGCUUGCCACUGAAUCCGAAAGGACAGGAGCAUACGGCCACGUCCUCCUGCUCACUGCAGCUU .(-----((.....--------...(((.......)))(((((((.(((((.....)))))..(((....))))))))))....))).........((((.....))))... ( -29.50) >DroEre_CAF1 1016 104 + 1 UGUCUGCCUCCCCA--------UUUGUAUUUGACUUACGCUCCUGGCAUUGCUUGCCACUGAAUCCGAAAGGACAGGAGCAUACGGCCAUGCCCUGCUGUUCGCUGCAGCUU .....(((......--------...(((.......)))((((((((((.....))))......(((....))).))))))....)))........(((((.....))))).. ( -34.10) >DroYak_CAF1 1188 99 + 1 UA-----CUCCGCA--------ACUGUAUUUGACUUACGCUCCUGACAGUGUUUUCCACUGAAUCCAACAGGACAGGCGCAUACGGCCACGUCCUCUUGUUCAGUGCAGAUU ..-----.......--------.(((((((.(((....((.((((.(((((.....)))))..(((....))))))).))..(((....)))......))).)))))))... ( -28.20) >DroAna_CAF1 905 104 + 1 AG-----CUCAUCA---GGAACUCUAAAGUAAACCUACGUUCUUGGCAGUGCUUGCCACUGAAGCCCACCAGGCAGGAGCACACAGCCUCCUCGUCCUGUUCGCUGCAGGUG .(-----((...((---(((((................)))))))(((((((((.......))))....(((((((((((.....).))))).).))))...))))).))). ( -27.89) >consensus UG_____CUCCCCA________UUUGUAUUUGACUUACGCUCCUGACAGUGCUUGCCACUGAAUCCGAAAGGACAGGAGCAUACGGCCACGUCCUCCUGUUCACUGCAGCUU ......................................(((((((.(((((.....)))))..(((....))))))))))................((((.....))))... (-22.70 = -22.62 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:56 2006