
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,750,525 – 20,750,617 |
| Length | 92 |
| Max. P | 0.821335 |

| Location | 20,750,525 – 20,750,617 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -16.31 |
| Energy contribution | -14.23 |
| Covariance contribution | -2.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
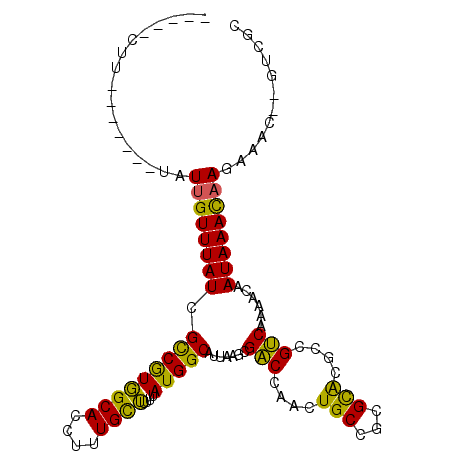
>X_DroMel_CAF1 20750525 92 + 22224390 -----GCC-------CAUAGUUUAUCGUCGUCGCACCUUUGCGUUUUAUGGCUUAAAGCGACCAACUGCCGCGUGCGGCGUCAAAACAAUAAACAAAAA----GUCGC -----...-------....((((((.((((((((((....((........)).....(((.........))))))))))).....)).)))))).....----..... ( -21.30) >DroVir_CAF1 91714 96 + 1 ACCCCCUU-------UAUUGUUUAUCGCCAUGGCACCUUUGUUUUUUAUGGCAUUAAGCGGCCAACUGCCGCGCA--CCGCCAAAACAAUAAAUAAGAAAC---UCGC ......((-------((((((((...((....))..............((((.....(((((.....)))))...--..))))))))))))))........---.... ( -25.70) >DroPse_CAF1 97911 96 + 1 -----CUU-------UAUUGUUUAUUGUCGUGGCACCUUUGCUUUUUAUGGCCCUAAGCGACCAACUGCCGCGCACGCCGUCAAAACAAUAAACAAGAAACUCGUGGC -----...-------..((((((((((((((((((...((((((...........)))))).....)))))))............)))))))))))............ ( -23.30) >DroYak_CAF1 57923 99 + 1 -----GCCCAUUGCCCAUUGUUUAUCGCCGUCGCACCUUUGCGUUUUAUGGCGUAAAGCGACCAACUGCCGCGUGCGGCGUCAAAACAAUAAACAAAAA----GUCGC -----.....(((...(((((((..(((((((((...((((((((....))))))))(((......))).))).))))))...)))))))...)))...----..... ( -28.80) >DroMoj_CAF1 134463 96 + 1 AUCCUCUU-------UAUUGUUUAUCGCCAUGGCACCUUUGUUUUUUAUGGCAUUAAGCGGCCAACUGCCGCGCA--CCGCCAAAACAAUAAAUAAGAAGC---UCGC .((...((-------((((((((...((....))..............((((.....(((((.....)))))...--..))))))))))))))...))...---.... ( -25.80) >DroPer_CAF1 99491 96 + 1 -----CUU-------UAUUGUUUAUUGUCGUGGCACCUUUGCUUUUUAUGGCCCUAAGCGACCAACUGCCGCGCACGCCGUCAAAACAAUAAACAAGAAACUCGUGGC -----...-------..((((((((((((((((((...((((((...........)))))).....)))))))............)))))))))))............ ( -23.30) >consensus _____CUU_______UAUUGUUUAUCGCCGUGGCACCUUUGCUUUUUAUGGCAUUAAGCGACCAACUGCCGCGCACGCCGUCAAAACAAUAAACAAGAAAC__GUCGC .................((((((((.(((((((((....))))....))))).......(((....(((...)))....)))......))))))))............ (-16.31 = -14.23 + -2.08)


| Location | 20,750,525 – 20,750,617 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20750525 92 - 22224390 GCGAC----UUUUUGUUUAUUGUUUUGACGCCGCACGCGGCAGUUGGUCGCUUUAAGCCAUAAAACGCAAAGGUGCGACGACGAUAAACUAUG-------GGC----- ....(----((...((((((((((..(((((((....)))).))).(((((.....((........))......)))))))))))))))...)-------)).----- ( -26.90) >DroVir_CAF1 91714 96 - 1 GCGA---GUUUCUUAUUUAUUGUUUUGGCGG--UGCGCGGCAGUUGGCCGCUUAAUGCCAUAAAAAACAAAGGUGCCAUGGCGAUAAACAAUA-------AAGGGGGU ....---...(((..(((((((((((((((.--((.(((((.....))))).)).))))).............(((....)))..))))))))-------))..))). ( -29.40) >DroPse_CAF1 97911 96 - 1 GCCACGAGUUUCUUGUUUAUUGUUUUGACGGCGUGCGCGGCAGUUGGUCGCUUAGGGCCAUAAAAAGCAAAGGUGCCACGACAAUAAACAAUA-------AAG----- ........(((.((((((((((((.((..(((.(..(((((.....)))))...).))).......(((....))))).)))))))))))).)-------)).----- ( -28.30) >DroYak_CAF1 57923 99 - 1 GCGAC----UUUUUGUUUAUUGUUUUGACGCCGCACGCGGCAGUUGGUCGCUUUACGCCAUAAAACGCAAAGGUGCGACGGCGAUAAACAAUGGGCAAUGGGC----- ....(----(..(((((((((((((...(((((...(((((.....)))))..............((((....)))).)))))..)))))))))))))..)).----- ( -34.70) >DroMoj_CAF1 134463 96 - 1 GCGA---GCUUCUUAUUUAUUGUUUUGGCGG--UGCGCGGCAGUUGGCCGCUUAAUGCCAUAAAAAACAAAGGUGCCAUGGCGAUAAACAAUA-------AAGAGGAU ....---...((((.(((((((((((((((.--((.(((((.....))))).)).))))).............(((....)))..))))))))-------)).)))). ( -29.70) >DroPer_CAF1 99491 96 - 1 GCCACGAGUUUCUUGUUUAUUGUUUUGACGGCGUGCGCGGCAGUUGGUCGCUUAGGGCCAUAAAAAGCAAAGGUGCCACGACAAUAAACAAUA-------AAG----- ........(((.((((((((((((.((..(((.(..(((((.....)))))...).))).......(((....))))).)))))))))))).)-------)).----- ( -28.30) >consensus GCGAC__GUUUCUUGUUUAUUGUUUUGACGGCGUGCGCGGCAGUUGGUCGCUUAAGGCCAUAAAAAGCAAAGGUGCCACGACGAUAAACAAUA_______AAG_____ ............(((((((((((..((.(.......(((((.....))))).....(((............)))).))..)))))))))))................. (-18.98 = -19.12 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:00 2006