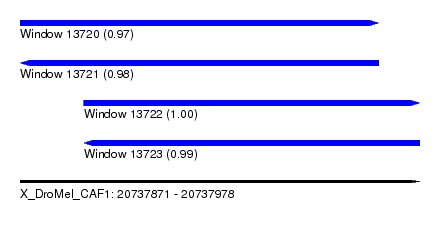
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,737,871 – 20,737,978 |
| Length | 107 |
| Max. P | 0.995164 |

| Location | 20,737,871 – 20,737,967 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -21.31 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
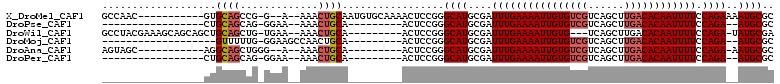
>X_DroMel_CAF1 20737871 96 + 22224390 GCCAAC-----------GUGCAGCCG-G--A--AAACUGCAAUGUGCAAAACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGC ((...(-----------((((..(((-(--(--....(((.....)))....))))))))))......(((((((((((((......)))))))))))))........)).. ( -33.10) >DroPse_CAF1 88034 81 + 1 -----------------CUGCAGCAG-GGAA--AAACUGCA---------ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA--AUGCGC -----------------.(((.((((-....--...)))).---------.(....)))).(((.((((((((((((((((......)))))))))))).))))--.))).. ( -27.00) >DroWil_CAF1 91451 96 + 1 GCCUACGAAAGCAGCAGCUGCAGCUG-UGAA--AAACUGCA---------ACUCCGGGCAUGCGAUUUGAAAAUUGUG---UCAGCUUGACACAAUUUUCCAGA-UAUGCGA ((((......((((((((....))))-....--...)))).---------.....)))).((((((((((((((((((---((.....))))))))))).))))-).)))). ( -33.71) >DroMoj_CAF1 120212 81 + 1 -------------------GUUUUUG-GGAAGCCAACUGCA---------ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA--AUGCGC -------------------((..(((-(....))))..)).---------...........(((.((((((((((((((((......)))))))))))).))))--.))).. ( -26.40) >DroAna_CAF1 1756 87 + 1 AGUAGC-----------AGGCAGCUGGG--A--AAACUGCA---------ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA-AAUGCGC ....((-----------(.((((((.((--(--........---------..))).))).))).....(((((((((((((......)))))))))))))....-..))).. ( -31.10) >DroPer_CAF1 90133 81 + 1 -----------------CUGCAGCAG-GGAA--AAACUGCA---------ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA--AUGCGC -----------------.(((.((((-....--...)))).---------.(....)))).(((.((((((((((((((((......)))))))))))).))))--.))).. ( -27.00) >consensus _________________CUGCAGCUG_GGAA__AAACUGCA_________ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA__AUGCGC ...................((((.............)))).................((((....((((((((((((((((......)))))))))))).))))..)))).. (-21.31 = -22.00 + 0.70)


| Location | 20,737,871 – 20,737,967 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -21.17 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20737871 96 - 22224390 GCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGUUUUGCACAUUGCAGUUU--U--C-CGGCUGCAC-----------GUUGGC (((.((((....((((((((((((........))))))))))))))))))).(((((((((..((((.....))))..)--)--)-)))..))).-----------...... ( -35.60) >DroPse_CAF1 88034 81 - 1 GCGCAU--UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU---------UGCAGUUU--UUCC-CUGCUGCAG----------------- (((.((--(...((((((((((((........)))))))))))).))))))...........---------((((((..--....-..)))))).----------------- ( -27.70) >DroWil_CAF1 91451 96 - 1 UCGCAUA-UCUGGAAAAUUGUGUCAAGCUGA---CACAAUUUUCAAAUCGCAUGCCCGGAGU---------UGCAGUUU--UUCA-CAGCUGCAGCUGCUGCUUUCGUAGGC ..((((.-....((((((((((((.....))---)))))))))).......))))..(.(((---------(((((((.--....-.)))))))))).).((((....)))) ( -35.20) >DroMoj_CAF1 120212 81 - 1 GCGCAU--UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU---------UGCAGUUGGCUUCC-CAAAAAC------------------- (((.((--((((((((((((((((........)))))))))))).....(....).))))))---------)))..((((....)-)))....------------------- ( -24.40) >DroAna_CAF1 1756 87 - 1 GCGCAUU-UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU---------UGCAGUUU--U--CCCAGCUGCCU-----------GCUACU (((.(((-(...((((((((((((........)))))))))))))))))))........(((---------.((((((.--.--...))))))..-----------)))... ( -27.60) >DroPer_CAF1 90133 81 - 1 GCGCAU--UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU---------UGCAGUUU--UUCC-CUGCUGCAG----------------- (((.((--(...((((((((((((........)))))))))))).))))))...........---------((((((..--....-..)))))).----------------- ( -27.70) >consensus GCGCAU__UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU_________UGCAGUUU__UUCC_CAGCUGCAG_________________ ..((((......((((((((((((........)))))))))))).......)))).................((((((.........))))))................... (-21.17 = -22.12 + 0.95)


| Location | 20,737,888 – 20,737,978 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.43 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.65 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20737888 90 + 22224390 -AAACUGCAAUGUGCAAAACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAG -....(((.....)))....(((((((..(((.((((((((((((((((......)))))))))))))...))).))))))))))...... ( -32.00) >DroVir_CAF1 78364 80 + 1 CCAACUGCA---------ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA--AUGCGCCUGGAACGUAG ....((((.---------..(((((((..(((.((((((((((((((((......)))))))))))).))))--.))))))))))..)))) ( -34.70) >DroGri_CAF1 84275 76 + 1 -CAAUUGCA---------ACUCCGGGCAUGCGAUUCGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA--AUGCGCCUGGAACG--- -........---------..(((((((..((.(((((((((((((((((......)))))))))))))..))--)))))))))))...--- ( -32.00) >DroWil_CAF1 91481 76 + 1 -AAACUGCA---------ACUCCGGGCAUGCGAUUUGAAAAUUGUG---UCAGCUUGACACAAUUUUCCAGA-UAUGCGACUGGAAAUGG- -......((---------..(((((...((((((((((((((((((---((.....))))))))))).))))-).)))).)))))..)).- ( -27.20) >DroMoj_CAF1 120224 80 + 1 CCAACUGCA---------ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA--AUGCGCCUGGAACGCAC .....(((.---------..(((((((..(((.((((((((((((((((......)))))))))))).))))--.))))))))))..))). ( -34.20) >DroAna_CAF1 1774 80 + 1 -AAACUGCA---------ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA-AAUGCGCCUGGAAUGGAA -........---------..(((((((..(((.((((((((((((((((......)))))))))))))..))-).))))))))))...... ( -30.70) >consensus _AAACUGCA_________ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGA__AUGCGCCUGGAACGGA_ ....................(((((((..(((.((((((((((((((((......)))))))))))).))))...))))))))))...... (-26.60 = -27.43 + 0.83)


| Location | 20,737,888 – 20,737,978 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20737888 90 - 22224390 CUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGUUUUGCACAUUGCAGUUU- ....(((((.((.((((......((((((((((((........)))))))))))).......))))))))))).((((.....))))...- ( -29.32) >DroVir_CAF1 78364 80 - 1 CUACGUUCCAGGCGCAU--UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU---------UGCAGUUGG .....((((.(((((((--(...((((((((((((........)))))))))))).))).))..))).)))).---------......... ( -25.50) >DroGri_CAF1 84275 76 - 1 ---CGUUCCAGGCGCAU--UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCGAAUCGCAUGCCCGGAGU---------UGCAAUUG- ---..((((.(((((((--((..((((((((((((........)))))))))))))))).))..))).)))).---------........- ( -27.40) >DroWil_CAF1 91481 76 - 1 -CCAUUUCCAGUCGCAUA-UCUGGAAAAUUGUGUCAAGCUGA---CACAAUUUUCAAAUCGCAUGCCCGGAGU---------UGCAGUUU- -............(((..-((((((((((((((((.....))---)))))))))).....(....).))))..---------))).....- ( -22.10) >DroMoj_CAF1 120224 80 - 1 GUGCGUUCCAGGCGCAU--UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU---------UGCAGUUGG .(((..(((.(((((((--(...((((((((((((........)))))))))))).))).))..))).)))..---------.)))..... ( -27.60) >DroAna_CAF1 1774 80 - 1 UUCCAUUCCAGGCGCAUU-UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU---------UGCAGUUU- ....(((((.((((....-....((((((((((((........))))))))))))........)))).)))))---------........- ( -26.69) >consensus _UCCGUUCCAGGCGCAU__UCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU_________UGCAGUUG_ .....((((.((.((((......((((((((((((........)))))))))))).......))))))))))................... (-23.30 = -23.80 + 0.50)

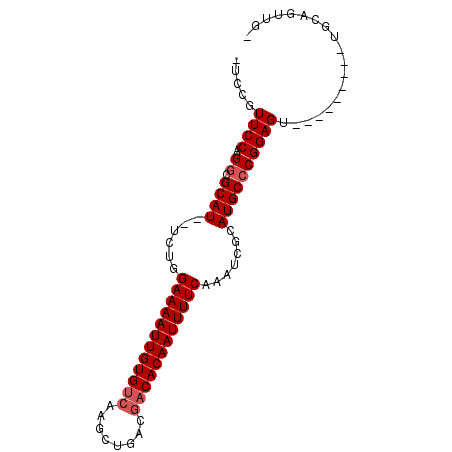
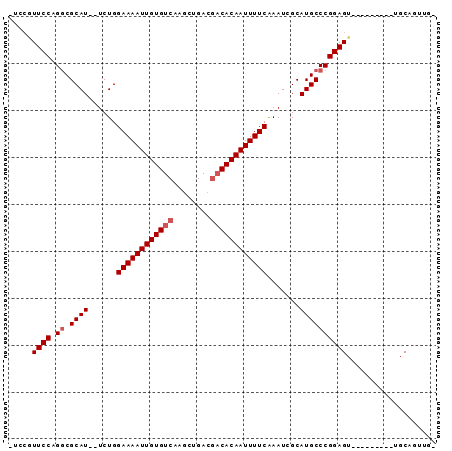
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:57 2006