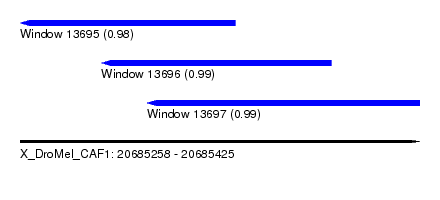
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,685,258 – 20,685,425 |
| Length | 167 |
| Max. P | 0.990575 |

| Location | 20,685,258 – 20,685,348 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 91.48 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -29.18 |
| Energy contribution | -28.63 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
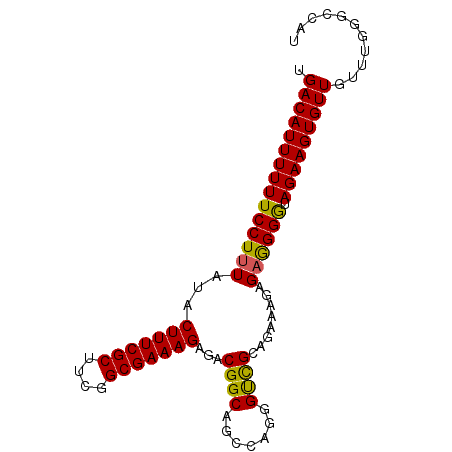
>X_DroMel_CAF1 20685258 90 - 22224390 UGACAUUUUUUCCUUUAUACUUUCGCUUCGGCGAAAGAGACGGCAGCCAGGGCCGCAGAAAGAGAAGGAUAGAAGUGUUGUUUGGGCCAU .(((((((((((((((...(((((((....))))))).(.((((.......))))).......)))))).)))))))))........... ( -32.80) >DroEre_CAF1 81645 89 - 1 UGACAUUUUUUCCUUUAUACUUUCGCUUCGGCGAAAGAGACGGCAGGCUGGGUCGCAGAAAAAG-GGGGUAGAAGUGUUGUUUGGGCCAU .(((((((((((((((...(((((((....))))))).(.((((.......)))))......))-)))).)))))))))........... ( -28.50) >DroYak_CAF1 83220 90 - 1 UGACAUUUUUUCCUUUAUACUUUCGCUUCGGCGAAAGAGACGGCAGCAGGGGUUGUGGAAAGAGAGGGGUAGAAGUGUUGUUUGGGCCAU .(((((((((((((((...(((((((....)))))))...(.(((((....))))).).....)))))).)))))))))........... ( -32.00) >consensus UGACAUUUUUUCCUUUAUACUUUCGCUUCGGCGAAAGAGACGGCAGCCAGGGUCGCAGAAAGAGAGGGGUAGAAGUGUUGUUUGGGCCAU .(((((((((((((((...(((((((....)))))))...((((.......))))........)))))).)))))))))........... (-29.18 = -28.63 + -0.55)


| Location | 20,685,292 – 20,685,388 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.50 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
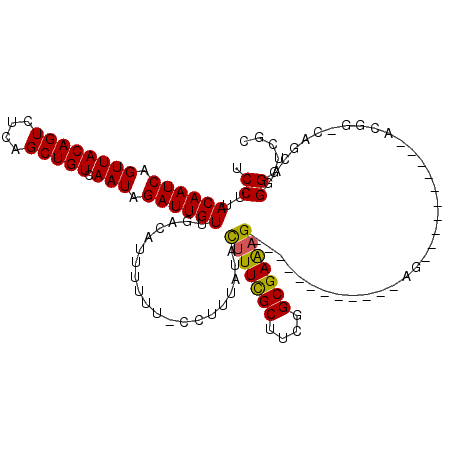
>X_DroMel_CAF1 20685292 96 - 22224390 UCCUUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUU-CCUUUAUACUUUCGCUUCGGCGAAAG------------AG----------ACGG-CAGCCAGGGCCGC .....((((((.((((((((....))))).))).))))))...........-........(((((((....)))))))------------..----------.(((-(.......)))). ( -29.10) >DroEre_CAF1 81678 96 - 1 UCCUUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUU-CCUUUAUACUUUCGCUUCGGCGAAAG------------AG----------ACGG-CAGGCUGGGUCGC ..................(.(((((((((((((.....)))))).......-........(((((((....)))))))------------..----------....-..))))))).).. ( -30.30) >DroYak_CAF1 83254 96 - 1 UCCUUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUU-CCUUUAUACUUUCGCUUCGGCGAAAG------------AG----------ACGG-CAGCAGGGGUUGU .....((((((.(((.(.(((((...(((((((.....)))))))......-.........((((((....)))))))------------))----------)).)-.)))...)))))) ( -30.00) >DroAna_CAF1 57358 114 - 1 GCCAUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUUUUCUCUUUUUUUUUG------CGAGUGUGUGUGUGUGGGAGUCCUCCUUUAAGGACCACCAGGGAGUGU ..((.((((((.((((((((....))))).))).))))))))((((((((...(((..........------.)))..........(((..(((((......)))))..))))))))))) ( -26.70) >consensus UCCUUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUU_CCUUUAUACUUUCGCUUCGGCGAAAG____________AG__________ACGG_CAGCAGGGGUCGC .((..((((((.((((((((....))))).))).))))))....................(((((((....)))))))...................................))..... (-20.75 = -20.50 + -0.25)


| Location | 20,685,311 – 20,685,425 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -21.74 |
| Consensus MFE | -19.30 |
| Energy contribution | -19.05 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20685311 114 - 22224390 CGCAUUUACUAGCCCC---GCCCGUUCCGUCUCUGUUUACUCCUUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUU-CCUUUAUACUUUCGCUUCGGCGAAAG-- .((........))...---.........(((..............((((((.((((((((....))))).))).)))))).))).......-........(((((((....)))))))-- ( -24.46) >DroEre_CAF1 81697 114 - 1 CGCAUUUCCCAGCCCC---GCCCAUUCCGCCUCUGUUUACUCCUUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUU-CCUUUAUACUUUCGCUUCGGCGAAAG-- .((........))...---..............((((........((((((.((((((((....))))).))).)))))).))))......-........(((((((....)))))))-- ( -23.50) >DroYak_CAF1 83273 117 - 1 CGCAUUUCCCAGCCCCUCUGCUUACUGCCCCUCUGUUUACUCCUUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUU-CCUUUAUACUUUCGCUUCGGCGAAAG-- .(((.....(((.....))).....))).....((((........((((((.((((((((....))))).))).)))))).))))......-........(((((((....)))))))-- ( -25.60) >DroAna_CAF1 57398 100 - 1 ----UUUAUCAUCCCC---ACCC-------CUUUUUUUUCGCCAUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUUUUCUCUUUUUUUUUG------CGAGUGUG ----............---....-------.......(((((((.((((((.((((((((....))))).))).)))))))).......................)------)))).... ( -13.40) >consensus CGCAUUUACCAGCCCC___GCCCAUUCCGCCUCUGUUUACUCCUUACAAUCAGUUACAGUCUCAGCUGUCAAUAGAUUGUUGACAUUUUUU_CCUUUAUACUUUCGCUUCGGCGAAAG__ .............................................((((((.((((((((....))))).))).))))))....................(((((((....))))))).. (-19.30 = -19.05 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:34 2006