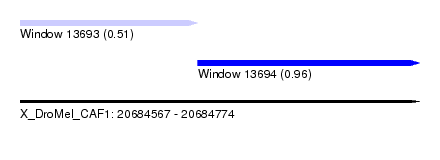
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,684,567 – 20,684,774 |
| Length | 207 |
| Max. P | 0.960738 |

| Location | 20,684,567 – 20,684,659 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 85.14 |
| Mean single sequence MFE | -16.67 |
| Consensus MFE | -11.63 |
| Energy contribution | -11.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20684567 92 + 22224390 CCGGCACGACGACAGACAUCUGUCGAUGGCUUCUCAACAGUCAUUUUCCGACGAAUACUAAAUUGCCUCUAAUCUACUUUAUCAUCGACACC ..((((...((((((....))))))((((((.......))))))...................))))......................... ( -18.70) >DroEre_CAF1 81030 76 + 1 ----------------CAUCUGUCGAUGGCUUCUCAACAGUCAUUUUCCGACGAAUACUAAAUUGCCUCUAAUCUACUCCAACAUCGACACC ----------------....((((((((...........(((.......)))((.............)).............)))))))).. ( -11.72) >DroYak_CAF1 82547 92 + 1 CCGGGACGACGACAGACAUCUGUCGAUGGCUUCUCAACAGUCAUUUUCCGACGAAUGCUAAAUUGCCUCUAAUCUACUUUAUCAUCGACACC .(((((...((((((....))))))((((((.......)))))).))))).(((..((......))...(((......)))...)))..... ( -19.60) >consensus CCGG_ACGACGACAGACAUCUGUCGAUGGCUUCUCAACAGUCAUUUUCCGACGAAUACUAAAUUGCCUCUAAUCUACUUUAUCAUCGACACC ....................((((((((...........(((.......))).(((.....)))..................)))))))).. (-11.63 = -11.63 + 0.00)



| Location | 20,684,659 – 20,684,774 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.41 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -13.17 |
| Energy contribution | -12.73 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20684659 115 + 22224390 AGGAAACAGACAGAGAGCCUAGGCAAGUGCUGGAGAAUGAGAAAGAGGCAAUUUCCAUUAACCACUUUGAAAAAUAAUGUAGUGUGGAUGAUG-AGAAAGUGUACAUUUUCAAAAC .(....)...(((...((....)).....)))(((((((........(((.(((((((((.((((..................)))).)))))-.)))).))).)))))))..... ( -24.77) >DroEre_CAF1 81106 105 + 1 AGGAAACAGACAGGGAGCCCCGGCGAAAGUUGGAGAAAAAGCCAGGGGCAAUUUCCAUUAACUAGUUUGAACAAUAAUGUAGUGGAGAUGAUA-AGAAAUGGGAAA---------- .(....).........((((((((....)))((........)).))))).((((((((((....(((........))).))))))))))....-............---------- ( -27.60) >DroYak_CAF1 82639 109 + 1 AGGAAAUAGACAGGGAGCCUCAGCGAAAGUUGCAGAAACAGUA-AGGGCAAUUUCCAUUAACCACUUUGAACAAUAAUGUUG-----AUGAUGGAGUAAGUGGAAAUU-AGAAAAC ................((((((((....))))...........-.))))((((((((((..(((...(.((((....)))).-----)...)))....))))))))))-....... ( -23.30) >consensus AGGAAACAGACAGGGAGCCUCGGCGAAAGUUGGAGAAAAAGAAAGGGGCAAUUUCCAUUAACCACUUUGAACAAUAAUGUAGUG__GAUGAUG_AGAAAGUGGAAAUU___AAAAC .((((((.....)...((((((((....)))).............))))..))))).....(((((((............................)))))))............. (-13.17 = -12.73 + -0.43)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:31 2006