
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,674,417 – 20,674,540 |
| Length | 123 |
| Max. P | 0.993921 |

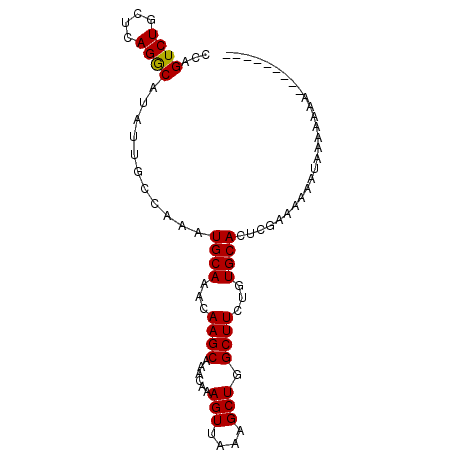
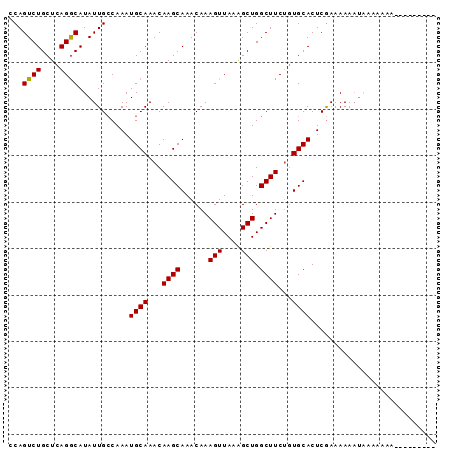
| Location | 20,674,417 – 20,674,508 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 83.15 |
| Mean single sequence MFE | -15.58 |
| Consensus MFE | -12.05 |
| Energy contribution | -11.83 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20674417 91 + 22224390 CCAGUCUGCUCAGGCAUAUUGGCAAAUGCAAACAAGCAAACAAAGUUAAGGCUGGCUUCUUUGCACUCAAAAAA-CAAAAAAAAAAAAAUAC (((((.(((....))).)))))....((((((.((((......(((....))).)))).)))))).........-................. ( -18.60) >DroEre_CAF1 70865 83 + 1 ACAGUCUGCUCAGGCAUAUUGCCAAAUGCAAACAAGCAAACAAAGUUAAAGCUGGCUUCUGUGCACUCGAAAAAAUGAAAUAU--------- ...((.(((...(((.....)))....))).))..(((....((((((....))))))...)))...................--------- ( -15.10) >DroYak_CAF1 72411 83 + 1 CCAGUCUGCUCAGACAUAUUGCCAAAUGCAAACAAGCAAACAAAGUUAAAGCUGGCUUCAGUGCACUCGAAAAAAUAUAAAAC--------- ...((((....))))(((((......((((...((((......(((....))).))))...))))........))))).....--------- ( -13.04) >consensus CCAGUCUGCUCAGGCAUAUUGCCAAAUGCAAACAAGCAAACAAAGUUAAAGCUGGCUUCUGUGCACUCGAAAAAAUAAAAAAA_________ ...((((....))))...........((((...((((......(((....))).))))...))))........................... (-12.05 = -11.83 + -0.22)

| Location | 20,674,417 – 20,674,508 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 83.15 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20674417 91 - 22224390 GUAUUUUUUUUUUUUUG-UUUUUUGAGUGCAAAGAAGCCAGCCUUAACUUUGUUUGCUUGUUUGCAUUUGCCAAUAUGCCUGAGCAGACUGG .................-......(((((((((.((((.(((.........))).)))).))))))))).(((...(((....)))...))) ( -20.50) >DroEre_CAF1 70865 83 - 1 ---------AUAUUUCAUUUUUUCGAGUGCACAGAAGCCAGCUUUAACUUUGUUUGCUUGUUUGCAUUUGGCAAUAUGCCUGAGCAGACUGU ---------..............((((((((.(.((((.(((.........))).)))).).))))))))(((...(((....)))...))) ( -19.50) >DroYak_CAF1 72411 83 - 1 ---------GUUUUAUAUUUUUUCGAGUGCACUGAAGCCAGCUUUAACUUUGUUUGCUUGUUUGCAUUUGGCAAUAUGUCUGAGCAGACUGG ---------((((.((((.((.(((((((((...((((.(((.........))).))))...))))))))).)).))))..))))....... ( -18.90) >consensus _________AUUUUUUAUUUUUUCGAGUGCACAGAAGCCAGCUUUAACUUUGUUUGCUUGUUUGCAUUUGGCAAUAUGCCUGAGCAGACUGG .......................((((((((.(.((((.(((.........))).)))).).))))))))(((...(((....)))...))) (-16.26 = -16.27 + 0.01)

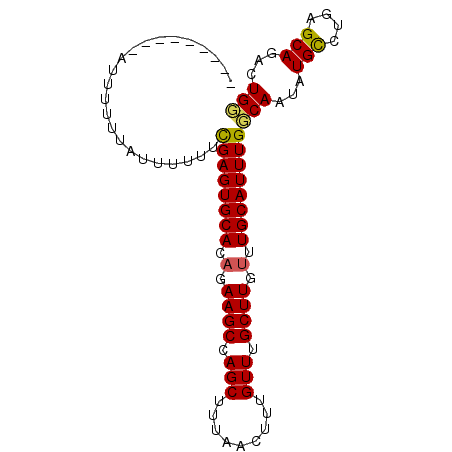
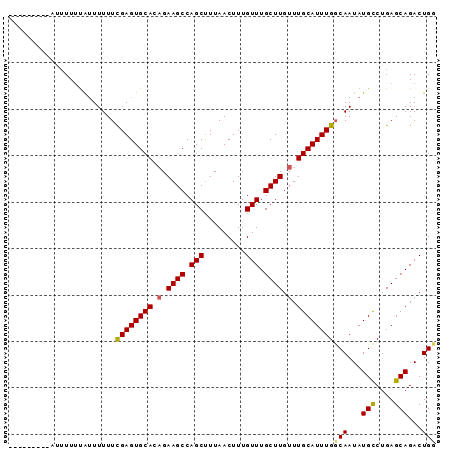
| Location | 20,674,434 – 20,674,540 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.75 |
| Mean single sequence MFE | -13.55 |
| Consensus MFE | -8.03 |
| Energy contribution | -8.03 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20674434 106 + 22224390 AUUGGCAAAUGCAAACAAGCAAACAAAGUUAAGGCUGGCUUCUUUGCACUCAAAAAA-CAAAAAAAAAAAAAUACAAGUAUCUGCGGUAUUUAUAAAUUUCAAAAAA---- .((((....((((((.((((......(((....))).)))).)))))).........-...........((((((..((....)).)))))).......))))....---- ( -14.10) >DroEre_CAF1 70882 91 + 1 AUUGCCAAAUGCAAACAAGCAAACAAAGUUAAAGCUGGCUUCUGUGCACUCGAAAAAAUGAAAUAU---------------AUCAGGUAUAACU-AAUUCC----AAAUUA ..((((...((((...((((......(((....))).))))...))))..........(((.....---------------.))))))).....-......----...... ( -12.80) >DroYak_CAF1 72428 96 + 1 AUUGCCAAAUGCAAACAAGCAAACAAAGUUAAAGCUGGCUUCAGUGCACUCGAAAAAAUAUAAAAC---------------CUGAAGUAUAACUUAAAUGCAGAAAAAUAA .((((....(((......)))....((((((......(((((((......................---------------))))))).))))))....))))........ ( -13.75) >consensus AUUGCCAAAUGCAAACAAGCAAACAAAGUUAAAGCUGGCUUCUGUGCACUCGAAAAAAUAAAAAAA_______________CUGAGGUAUAACU_AAUUCCA_AAAAAU_A .........((((...((((......(((....))).))))...))))............................................................... ( -8.03 = -8.03 + -0.00)


| Location | 20,674,434 – 20,674,540 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.75 |
| Mean single sequence MFE | -17.97 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20674434 106 - 22224390 ----UUUUUUGAAAUUUAUAAAUACCGCAGAUACUUGUAUUUUUUUUUUUUUG-UUUUUUGAGUGCAAAGAAGCCAGCCUUAACUUUGUUUGCUUGUUUGCAUUUGCCAAU ----......................(((((((....)))))...........-......(((((((((.((((.(((.........))).)))).))))))))))).... ( -18.00) >DroEre_CAF1 70882 91 - 1 UAAUUU----GGAAUU-AGUUAUACCUGAU---------------AUAUUUCAUUUUUUCGAGUGCACAGAAGCCAGCUUUAACUUUGUUUGCUUGUUUGCAUUUGGCAAU .....(----(((((.-.((((....))))---------------..)))))).....(((((((((.(.((((.(((.........))).)))).).))))))))).... ( -19.40) >DroYak_CAF1 72428 96 - 1 UUAUUUUUCUGCAUUUAAGUUAUACUUCAG---------------GUUUUAUAUUUUUUCGAGUGCACUGAAGCCAGCUUUAACUUUGUUUGCUUGUUUGCAUUUGGCAAU .........((((((((((.....))).))---------------))............((((((((...((((.(((.........))).))))...))))))))))).. ( -16.50) >consensus U_AUUUUU_UGAAAUU_AGUUAUACCUCAG_______________AUUUUUUAUUUUUUCGAGUGCACAGAAGCCAGCUUUAACUUUGUUUGCUUGUUUGCAUUUGGCAAU ...........................................................((((((((.(.((((.(((.........))).)))).).))))))))..... (-14.22 = -14.33 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:19 2006