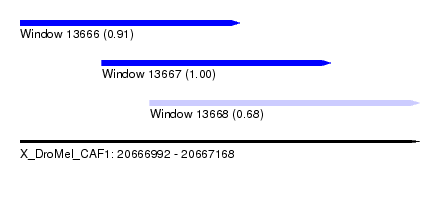
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,666,992 – 20,667,168 |
| Length | 176 |
| Max. P | 0.996881 |

| Location | 20,666,992 – 20,667,089 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.26 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20666992 97 + 22224390 --CCCCAUCCUGCCACAGGCCUAGCCCAAGCCAGCCCC--------------------AUCCCCUGGAA-AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUU --........(((....(((...((....))..)))..--------------------.......((((-.(((((.....)))))...)))))))((((((....))))))........ ( -23.30) >DroSec_CAF1 68643 96 + 1 --CCCCGUCCCGCCACAGGCCUAGCCCAAGCAA-CCCC--------------------ACCCCCUGGAA-AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUU --.........((....(((...))).......-....--------------------.......((((-.(((((.....)))))...)))))).((((((....))))))........ ( -20.90) >DroSim_CAF1 46122 97 + 1 --CCCCGUCCCGCCACAGGCCUAGCCCAAGCAAGCCCC--------------------ACCCCCUGGAA-AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUU --.........((....(((...((....))..)))..--------------------.......((((-.(((((.....)))))...)))))).((((((....))))))........ ( -23.10) >DroEre_CAF1 64317 117 + 1 --CCCCCUCCCCCCACAGGCGUAGCCCAAGCAAGCGCCACAGCACCACAGCACCCCCACCCCCCUGGAA-AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUU --...............(((((.((....))..)))))...((......))..............((((-.(((((.....)))))...))))...((((((....))))))........ ( -27.50) >DroYak_CAF1 65766 105 + 1 ACCCCCCUCCCACCGCAGGCCUAGCCCAAGCAAGCACC---------------UCCCUCCCCCCUGGAAAAACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUU ..............(((((.((.((....)).))..))---------------)...........((((..(((((.....)))))...)))))).((((((....))))))........ ( -23.10) >consensus __CCCCCUCCCGCCACAGGCCUAGCCCAAGCAAGCCCC____________________ACCCCCUGGAA_AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUU ..................((...((....))..................................((((..(((((.....)))))...)))))).((((((....))))))........ (-20.26 = -20.26 + -0.00)


| Location | 20,667,028 – 20,667,129 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20667028 101 + 22224390 ------------------AUCCCCUGGAA-AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCU ------------------.......((((-.(((((.....)))))...))))((((...(((..((((((((.....)))))))).)))...((((....))))))))........... ( -23.50) >DroSec_CAF1 68678 101 + 1 ------------------ACCCCCUGGAA-AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCU ------------------.......((((-.(((((.....)))))...))))((((...(((..((((((((.....)))))))).)))...((((....))))))))........... ( -23.50) >DroSim_CAF1 46158 101 + 1 ------------------ACCCCCUGGAA-AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCU ------------------.......((((-.(((((.....)))))...))))((((...(((..((((((((.....)))))))).)))...((((....))))))))........... ( -23.50) >DroEre_CAF1 64355 119 + 1 AGCACCACAGCACCCCCACCCCCCUGGAA-AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCU .((......)).....((((((.((((..-......)))).)))))).....(((((...(((..((((((((.....)))))))).)))...((((....))))))))).......... ( -27.10) >DroYak_CAF1 65804 107 + 1 -------------UCCCUCCCCCCUGGAAAAACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCU -------------............((((..(((((.....)))))...))))((((...(((..((((((((.....)))))))).)))...((((....))))))))........... ( -23.90) >consensus __________________ACCCCCUGGAA_AACCCCUUAGCGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCU .........................((((..(((((.....)))))...))))((((...(((..((((((((.....)))))))).)))...((((....))))))))........... (-24.46 = -24.46 + 0.00)



| Location | 20,667,049 – 20,667,168 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.66 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -27.13 |
| Energy contribution | -27.13 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20667049 119 + 22224390 CGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCUUCCUACGUUUUGCCUU-UUUAAAGUGGCGCCCCCAACGGC .((((((.........((((((....))))))..(((((((((.((((.(((.((.((......(((....)))......)).)).))).))))..-))).))))))))))))....... ( -28.20) >DroSec_CAF1 68699 119 + 1 CGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCUUCCUUCGUUUUGCCUU-UUUAAAGUGGCGCCCCCAACGGC .((((((((.....((.((((..(.((((((((.....)))))))).)..)))).))((((((.((.(..((................))..))).-..))))))))))))))....... ( -27.89) >DroSim_CAF1 46179 119 + 1 CGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCUUCCUUCGUUUUGCCUU-UUUAAAGUGGCGCCCCCAACGGC .((((((((.....((.((((..(.((((((((.....)))))))).)..)))).))((((((.((.(..((................))..))).-..))))))))))))))....... ( -27.89) >DroEre_CAF1 64394 118 + 1 CGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCUUCCUUCGCUUUGCCUU--UUAAAGUGGCGCCCCCAACGGC .((((((((...(((((...(((..((((((((.....)))))))).)))...((((....)))))))))................((((((....--.))))))))))))))....... ( -30.60) >DroYak_CAF1 65831 120 + 1 CGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCUUCCUUCGUUUUGCCUUUUUUAAAGUGGCGCCCCCAACGGC .((((((((.....((.((((..(.((((((((.....)))))))).)..)))).))((((((.((.(..((................))..)))....))))))))))))))....... ( -27.29) >consensus CGGGGUGUCUUCCGCACAAAUGCCUCCAUUUGACCCACUUAAAUGGCGUAAUUUAUGAUUUUAUAGUGUGAAUUUUUUCUUCCUUCGUUUUGCCUU_UUUAAAGUGGCGCCCCCAACGGC .((((((((.....((.((((..(.((((((((.....)))))))).)..)))).))((((((.((.(..((................))..)))....))))))))))))))....... (-27.13 = -27.13 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:08 2006