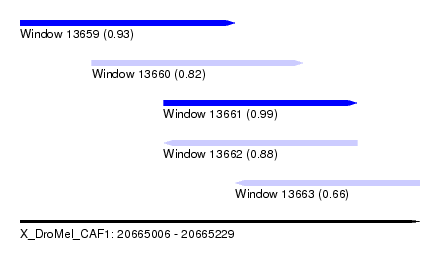
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,665,006 – 20,665,229 |
| Length | 223 |
| Max. P | 0.992788 |

| Location | 20,665,006 – 20,665,126 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -21.46 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20665006 120 + 22224390 GCAUACAUAUAUCAUUUGCAUAGUUAGACAAAGAUGGCCGAUAAGUCGAUGCGAAAUUUAAUCCGCAUUAAACGCUUCUAACAAAUGCACUAAAUAAAAAUCGAAAUAAAUUGCUUAAUA (((....(((.((((((((((.((((((.......(((......)))((((((..........)))))).......))))))..))))).........))).)).)))...)))...... ( -18.00) >DroSec_CAF1 66649 116 + 1 GCA----UAUAUCAUUUGCAUAGUUAGACAAAGAUGGCCGAUAAGUCGAUGCGAAAUUUAAUCUGCAUUAAACGCUUCUAACAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUA ...----..((((...(((((.((((((.......(((......)))((((((..........)))))).......))))))..))))).........((((((......)))))))))) ( -21.50) >DroSim_CAF1 44112 116 + 1 GCA----UAUAUCAUUUGCAUAGUUAGACAAAGAUGGCCGAUAAGUCGAUGCGAAAUUUAAUCCGCAUUAAACGCUUCUAACAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUA ...----..((((...(((((.((((((.......(((......)))((((((..........)))))).......))))))..))))).........((((((......)))))))))) ( -23.50) >DroEre_CAF1 62328 116 + 1 GCA----UAUAUCAUUUGCAUAGUUAGACAAAGAUGGUUGAUAAGUCGAUGCGAAAUUUAAGCCGCAUUAAACGCUUCUAGCAAAUGCACUAAAUAAAAUGCGAAAUAAAUUGCGUGAUA ...----..((((((.(((((.((((((....(((.........)))((((((..........)))))).......))))))..)))))...........((((......)))))))))) ( -22.20) >DroYak_CAF1 63645 116 + 1 GCA----UAUAUCAUUUGCAUAGUUAGACAAAGAUGGCCGAUAAGUCGAUGCGAAAUUAAAGCCGCAUCAAACGCUUCUACCAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUA ...----..((((...(((((...((((.......(((......)))((((((..........)))))).......))))....))))).........((((((......)))))))))) ( -22.10) >consensus GCA____UAUAUCAUUUGCAUAGUUAGACAAAGAUGGCCGAUAAGUCGAUGCGAAAUUUAAUCCGCAUUAAACGCUUCUAACAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUA .........((((...(((((.((((((.......(((......)))((((((..........)))))).......))))))..))))).........((((((......)))))))))) (-20.42 = -20.10 + -0.32)


| Location | 20,665,046 – 20,665,164 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.53 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20665046 118 + 22224390 AUAAGUCGAUGCGAAAUUUAAUCCGCAUUAAACGCUUCUAACAAAUGCACUAAAUAAAAAUCGAAAUAAAUUGCUUAAUAGUGAAAGUGCUUUGGAUGGCUAAUUUAACUGAAUAGCU ....(((((((((..........))))))............((((.(((((........((....))...(..((....))..).))))))))))))(((((.((.....)).))))) ( -18.30) >DroSec_CAF1 66685 116 + 1 AUAAGUCGAUGCGAAAUUUAAUCUGCAUUAAACGCUUCUAACAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUAGUGGAAGUGCUUUGGAUGGGUAAUUUAACUGAAUGG-- ((..(((((.(((...((((((....))))))))).))...((((.(((((...((..((((((......))))))..)).....))))))))))))..))...............-- ( -20.10) >DroSim_CAF1 44148 118 + 1 AUAAGUCGAUGCGAAAUUUAAUCCGCAUUAAACGCUUCUAACAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUAGUGGAAGUGCUUUGGAUGGCUAAUUUAACUGAAUGGCU ...((((((((((..........))))))....(((.....((((.(((((...((..((((((......))))))..)).....)))))))))...)))..............)))) ( -25.60) >DroEre_CAF1 62364 113 + 1 AUAAGUCGAUGCGAAAUUUAAGCCGCAUUAAACGCUUCUAGCAAAUGCACUAAAUAAAAUGCGAAAUAAAUUGCGUGAUAGUGGAAGUGCUUUGGACGGCUAAUUUAACUGAA----- ...((((((((((..........)))))((((((((((((((....))..........((((((......)))))).....)))))))).))))..)))))............----- ( -25.80) >DroYak_CAF1 63681 118 + 1 AUAAGUCGAUGCGAAAUUAAAGCCGCAUCAAACGCUUCUACCAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUAGUGGAAGUGCUUUGGAUGGCUAAUUCAACAGAAUGGCU ...((((((((((..........))))))....(((....(((((.(((((...((..((((((......))))))..)).....))))))))))..)))..((((....)))))))) ( -31.10) >consensus AUAAGUCGAUGCGAAAUUUAAUCCGCAUUAAACGCUUCUAACAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUAGUGGAAGUGCUUUGGAUGGCUAAUUUAACUGAAUGGCU ...((((((((((..........))))))............((((.(((((...((..((((((......))))))..)).....)))))))))...))))................. (-21.34 = -21.10 + -0.24)

| Location | 20,665,086 – 20,665,194 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -14.66 |
| Energy contribution | -16.62 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20665086 108 + 22224390 ACAAAUGCACUAAAUAAAAAUCGAAAUAAAUUGCUUAAUAGUGAAAGUGCUUUGGAUGGCUAAUUUAACUGAAUAGCUAAAU----AAAAGCCAUUCAUAAAA--AAAACUAAA ......(((((........((....))...(..((....))..).)))))..(((((((((.(((((.((....)).)))))----...))))))))).....--......... ( -17.80) >DroSec_CAF1 66725 102 + 1 ACAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUAGUGGAAGUGCUUUGGAUGGGUAAUUUAACUGAAUGG-----A----AUAAGCCAUUCAUAAAA--AAAA-CAAA .((((.(((((...((..((((((......))))))..)).....)))))))))..((..(..((((..(((((((-----.----.....))))))))))).--.)..-)).. ( -21.90) >DroSim_CAF1 44188 109 + 1 ACAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUAGUGGAAGUGCUUUGGAUGGCUAAUUUAACUGAAUGGCUAAAU----AUAAGCCAUUCAUAAAAAAAAAA-CAAA .((((.(((((...((..((((((......))))))..)).....)))))))))...............(((((((((....----...)))))))))...........-.... ( -26.00) >DroEre_CAF1 62404 96 + 1 GCAAAUGCACUAAAUAAAAUGCGAAAUAAAUUGCGUGAUAGUGGAAGUGCUUUGGACGGCUAAUUUAACUGAA--------------UAUGGCAAUC-UGAAA--AUAG-CAAA ((....(((((...((..((((((......))))))..)).....)))))((..((..((((((((....)))--------------).))))..))-..)).--...)-)... ( -21.40) >DroYak_CAF1 63721 110 + 1 CCAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUAGUGGAAGUGCUUUGGAUGGCUAAUUCAACAGAAUGGCUAAAUAUGUAUAAGCCGAAC-UAAAC--AUAA-CAGA (((((.(((((...((..((((((......))))))..)).....)))))))))).((((((.(((....)))))))))..(((((.((........-)).))--))).-.... ( -24.90) >consensus ACAAAUGCACUAAAUAAAAAGCGAAAUAAAUUGCUUGAUAGUGGAAGUGCUUUGGAUGGCUAAUUUAACUGAAUGGCUAAAU____AUAAGCCAUUCAUAAAA__AAAA_CAAA .((((.(((((...((..((((((......))))))..)).....)))))))))................((((((((...........))))))))................. (-14.66 = -16.62 + 1.96)

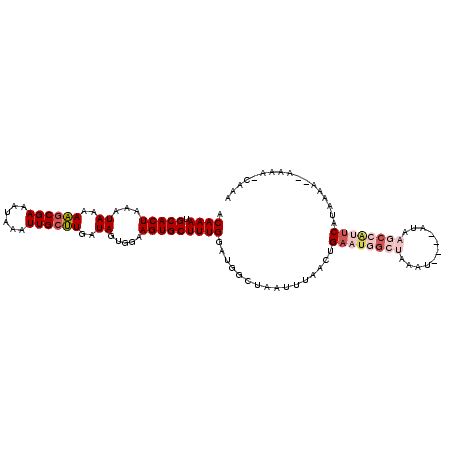

| Location | 20,665,086 – 20,665,194 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -19.25 |
| Consensus MFE | -11.34 |
| Energy contribution | -13.82 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20665086 108 - 22224390 UUUAGUUUU--UUUUAUGAAUGGCUUUU----AUUUAGCUAUUCAGUUAAAUUAGCCAUCCAAAGCACUUUCACUAUUAAGCAAUUUAUUUCGAUUUUUAUUUAGUGCAUUUGU .........--.....((.((((((...----((((((((....)))))))).)))))).))..(((..(.(((((.((((.((((......))))))))..))))).)..))) ( -20.20) >DroSec_CAF1 66725 102 - 1 UUUG-UUUU--UUUUAUGAAUGGCUUAU----U-----CCAUUCAGUUAAAUUACCCAUCCAAAGCACUUCCACUAUCAAGCAAUUUAUUUCGCUUUUUAUUUAGUGCAUUUGU ...(-(..(--((...(((((((.....----.-----)))))))...)))..)).....(((((((((.....((..((((..........))))..))...))))).)))). ( -19.30) >DroSim_CAF1 44188 109 - 1 UUUG-UUUUUUUUUUAUGAAUGGCUUAU----AUUUAGCCAUUCAGUUAAAUUAGCCAUCCAAAGCACUUCCACUAUCAAGCAAUUUAUUUCGCUUUUUAUUUAGUGCAUUUGU ...(-((..(((....(((((((((...----....)))))))))...)))..)))....(((((((((.....((..((((..........))))..))...))))).)))). ( -24.80) >DroEre_CAF1 62404 96 - 1 UUUG-CUAU--UUUCA-GAUUGCCAUA--------------UUCAGUUAAAUUAGCCGUCCAAAGCACUUCCACUAUCACGCAAUUUAUUUCGCAUUUUAUUUAGUGCAUUUGC ...(-(((.--(((..-(((((.....--------------..)))))))).))))....(((((((((..........((.((.....))))..........))))).)))). ( -11.25) >DroYak_CAF1 63721 110 - 1 UCUG-UUAU--GUUUA-GUUCGGCUUAUACAUAUUUAGCCAUUCUGUUGAAUUAGCCAUCCAAAGCACUUCCACUAUCAAGCAAUUUAUUUCGCUUUUUAUUUAGUGCAUUUGG ....-....--.....-....((((.......(((((((......))))))).))))..((((((((((.....((..((((..........))))..))...))))).))))) ( -20.70) >consensus UUUG_UUUU__UUUUAUGAAUGGCUUAU____AUUUAGCCAUUCAGUUAAAUUAGCCAUCCAAAGCACUUCCACUAUCAAGCAAUUUAUUUCGCUUUUUAUUUAGUGCAUUUGU .................((((((((...........))))))))................(((((((((.....((..((((..........))))..))...))))).)))). (-11.34 = -13.82 + 2.48)

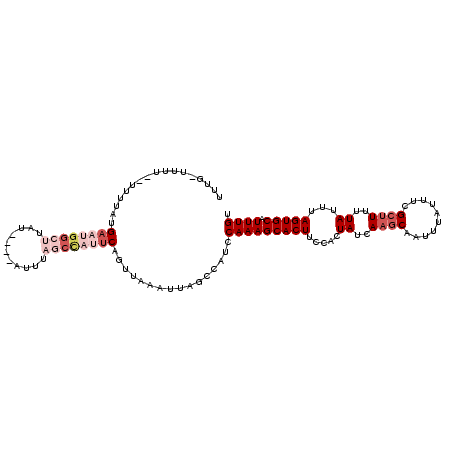
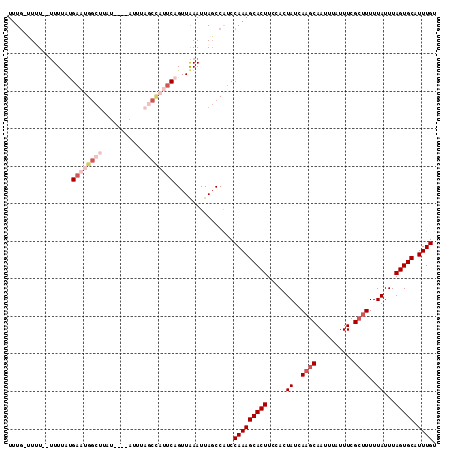
| Location | 20,665,126 – 20,665,229 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.22 |
| Mean single sequence MFE | -18.04 |
| Consensus MFE | -5.97 |
| Energy contribution | -9.16 |
| Covariance contribution | 3.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20665126 103 - 22224390 UGUUAGCAUUUCAU-----CUAAGCCAUUAGCGGCUUGUUUUUAGUUUU--UUUUAUGAAUGGCUUUU----AUUUAGCUAUUCAGUUAAAUUAGCCAUCCAAAGCACUUUCAC .....((.......-----.((((((......))))))...........--.....((.((((((...----((((((((....)))))))).)))))).))..))........ ( -24.60) >DroSec_CAF1 66765 97 - 1 UGUUGGCAUUUCAU-----CUAAGCCACUAGCGGUUUGAUUUUG-UUUU--UUUUAUGAAUGGCUUAU----U-----CCAUUCAGUUAAAUUACCCAUCCAAAGCACUUCCAC ..((((......((-----(.(((((......))))))))...(-(..(--((...(((((((.....----.-----)))))))...)))..))....))))........... ( -17.20) >DroSim_CAF1 44228 104 - 1 UGUUGGCAUUUCAU-----CUAAGCCGUUAGCGGUUUGAUUUUG-UUUUUUUUUUAUGAAUGGCUUAU----AUUUAGCCAUUCAGUUAAAUUAGCCAUCCAAAGCACUUCCAC ...((((.....((-----(.((((((....)))))))))....-...........(((((((((...----....))))))))).........))))................ ( -27.30) >DroEre_CAF1 62444 96 - 1 UUUGAGCAACCCAUAAAUUCUUACCCAUUAACAAUUUGUUUUUG-CUAU--UUUCA-GAUUGCCAUA--------------UUCAGUUAAAUUAGCCGUCCAAAGCACUUCCAC (((((((((...(((((((.(((.....))).)))))))..)))-))..--.....-(((.((...(--------------((......)))..)).))))))).......... ( -9.30) >DroYak_CAF1 63761 96 - 1 UGUUAGCAUCCCAUCAAUUCUAA--------------GUUUCUG-UUAU--GUUUA-GUUCGGCUUAUACAUAUUUAGCCAUUCUGUUGAAUUAGCCAUCCAAAGCACUUCCAC (((((((((..((..(((.....--------------)))..))-..))--)))..-....((((.......(((((((......))))))).))))......))))....... ( -11.80) >consensus UGUUAGCAUUUCAU_____CUAAGCCAUUAGCGGUUUGUUUUUG_UUUU__UUUUAUGAAUGGCUUAU____AUUUAGCCAUUCAGUUAAAUUAGCCAUCCAAAGCACUUCCAC ..(((((.............((((((......))))))...................((((((((...........)))))))).)))))........................ ( -5.97 = -9.16 + 3.19)

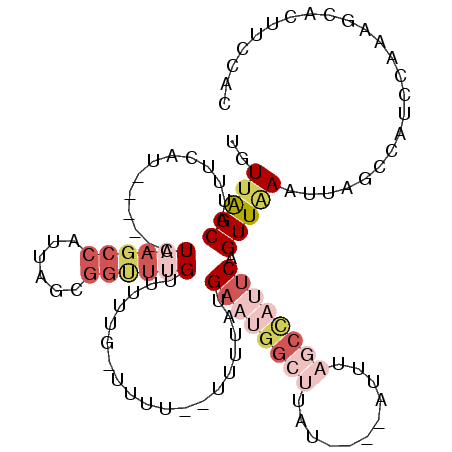

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:03 2006