
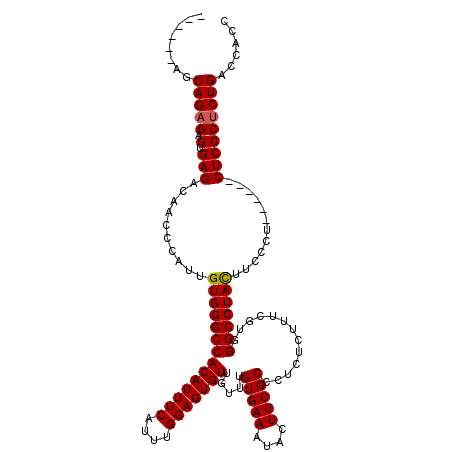
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,664,681 – 20,664,789 |
| Length | 108 |
| Max. P | 0.965408 |

| Location | 20,664,681 – 20,664,789 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.89 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -29.13 |
| Energy contribution | -29.17 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20664681 108 + 22224390 ------AGCAGUGAGUGAGACAACCCAUUGUGGGCCACAUUUCAUUUGGAGUGUUGUUUGUGAAAUACUUCGCCCUCUCUUUCGUGGGCCUACUUCCCU------CUCCCUCUGACCAUC ------......(((.((((.........((((((((((((((....))))))).....(((((....))))).............))))))).....)------))).)))........ ( -27.74) >DroSec_CAF1 66324 108 + 1 ------AGCAGAGAGUGAGACAACCCAUUGUGGGCCACAUUUCAUUUGGAGUGUUGUUUGUGAAAUACUUCGCCCUCUCUUUCGUGGGCCUACUUCCCU------CUCCCUCUGACCACC ------..(((((.(.((((.........((((((((((((((....))))))).....(((((....))))).............))))))).....)------)))))))))...... ( -31.54) >DroSim_CAF1 43783 108 + 1 ------AGCAGAGAGUGAGACAACCCAUUGUGGGCCACAUUUCAUUUGGAGUGUUGUUUGUGAAAUACUUCGCCAUCUCUUUCGUGGGCCUACUUCCCU------CUCCCUCUGACCACC ------..(((((.(.((((.........((((((((((((((....))))))).....(((((....))))).............))))))).....)------)))))))))...... ( -31.54) >DroEre_CAF1 61991 114 + 1 ------AGCAGAGAGUGAGGAAAACCAUUGUGGGCCACAUUUCAUUUGGAGUGUAGUUUGUGAAAUAUUUCGCCCUCUUUUUCGUGGGCCUAUCUCCCUCUCACUCUCCCUCUGACCGCC ------....(((((((((((((....(..(..((.(((((((....))))))).))..)..)....))))((((.(......).))))..........)))))))))............ ( -36.00) >DroYak_CAF1 63305 114 + 1 GCAGAGAGCAGAGAGUGAGGAAAGCCAUUGUGGGCCACAUUUCAUUUGGAGUGUAGAUUGUGAAAUAUUUCGCCCUCUCUUUCGUGGGCCUACUUCCCU------CUCCCUCUGACCAGC .(((((.(..(((((.((((....))...((((((((((((((....)))))))(((..(((((....)))))..)))........))))))).)).))------)))))))))...... ( -38.20) >consensus ______AGCAGAGAGUGAGACAACCCAUUGUGGGCCACAUUUCAUUUGGAGUGUUGUUUGUGAAAUACUUCGCCCUCUCUUUCGUGGGCCUACUUCCCU______CUCCCUCUGACCACC ........(((((.(.(((..........((((((((((((((....))))))).....(((((....))))).............)))))))............)))))))))...... (-29.13 = -29.17 + 0.04)


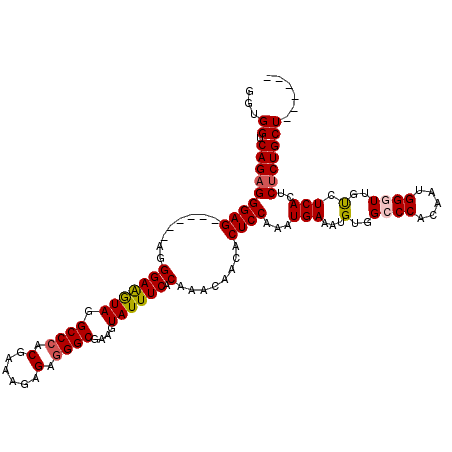
| Location | 20,664,681 – 20,664,789 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.89 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -26.97 |
| Energy contribution | -27.21 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20664681 108 - 22224390 GAUGGUCAGAGGGAG------AGGGAAGUAGGCCCACGAAAGAGAGGGCGAAGUAUUUCACAAACAACACUCCAAAUGAAAUGUGGCCCACAAUGGGUUGUCUCACUCACUGCU------ ...((.(((...(((------.((((....(((((((....)...((((.(..(((((((................)))))))).))))....)))))).)))).))).)))))------ ( -30.79) >DroSec_CAF1 66324 108 - 1 GGUGGUCAGAGGGAG------AGGGAAGUAGGCCCACGAAAGAGAGGGCGAAGUAUUUCACAAACAACACUCCAAAUGAAAUGUGGCCCACAAUGGGUUGUCUCACUCUCUGCU------ ...((.(((((((.(------(((((.((..((((.(....)...))))(((....))).........))))).........(..((((.....))))..)))).)))))))))------ ( -34.10) >DroSim_CAF1 43783 108 - 1 GGUGGUCAGAGGGAG------AGGGAAGUAGGCCCACGAAAGAGAUGGCGAAGUAUUUCACAAACAACACUCCAAAUGAAAUGUGGCCCACAAUGGGUUGUCUCACUCUCUGCU------ ...((.(((((((.(------(((((.((..(((..(....)....)))(((....))).........))))).........(..((((.....))))..)))).)))))))))------ ( -30.50) >DroEre_CAF1 61991 114 - 1 GGCGGUCAGAGGGAGAGUGAGAGGGAGAUAGGCCCACGAAAAAGAGGGCGAAAUAUUUCACAAACUACACUCCAAAUGAAAUGUGGCCCACAAUGGUUUUCCUCACUCUCUGCU------ ...((.(((((((.(((.(((((((((.(((((((.(......).))))(((....))).....)))..))))........((((...))))....)))))))).)))))))))------ ( -34.80) >DroYak_CAF1 63305 114 - 1 GCUGGUCAGAGGGAG------AGGGAAGUAGGCCCACGAAAGAGAGGGCGAAAUAUUUCACAAUCUACACUCCAAAUGAAAUGUGGCCCACAAUGGCUUUCCUCACUCUCUGCUCUCUGC ((.(..(((((((.(------((((((((.......(....)...((((...((((((((................)))))))).))))......))))))))).)))))))..)...)) ( -40.29) >consensus GGUGGUCAGAGGGAG______AGGGAAGUAGGCCCACGAAAGAGAGGGCGAAGUAUUUCACAAACAACACUCCAAAUGAAAUGUGGCCCACAAUGGGUUGUCUCACUCUCUGCU______ ...((.(((((((((........(((((((.((((.(......).))))....)))))).)........))))...(((...(..((((.....))))..).)))..)))))))...... (-26.97 = -27.21 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:58 2006