
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,663,911 – 20,664,085 |
| Length | 174 |
| Max. P | 0.962162 |

| Location | 20,663,911 – 20,664,022 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -27.47 |
| Energy contribution | -28.55 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
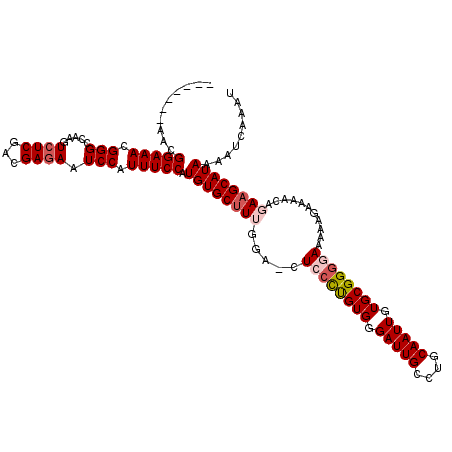
>X_DroMel_CAF1 20663911 111 - 22224390 -------AACGGAAACGGGCCAAGUCUCGACGAAAAUCCAUUUCCAUGUGCUUUGGA-CUCGUUGUGGGAUUGCCUGCAAUUGUGCGGGAAAAAGGAAAACAGAAGCAUAAAAUCAAAU -------...(((((.(((....((....)).....))).))))).((((((((...-(((((((..((....))..)).....))))).....(.....).))))))))......... ( -26.50) >DroSec_CAF1 65587 111 - 1 -------AGCGGAAACGGGCCAAGUCUCGACGAGAAUCCAUUUCCAUGUGCUUUGGA-CUCCCUGUGGGAUUGCCUGCAAUUGUGCGGGGAAAAAGAAAACAGAAGCAUAAAAUCAAAU -------.(((((((.(((.....((((...)))).))).))))).(((.((((...-.((((((((.(((((....))))).)))))))).))))...)))...))............ ( -34.00) >DroSim_CAF1 43045 111 - 1 -------AGCGGAAACGGGCCAAGUCUCGACGAGAAUCCAUUUCCAUGUGCUUUGGA-CUCCCUGUGUGAUUGCCUGCAAUUGUGCGGGGAAAAAGAAAACAGAAGCAUAAAAUCAAAU -------.(((((((.(((.....((((...)))).))).))))).(((.((((...-.((((((((..((((....))))..)))))))).))))...)))...))............ ( -34.80) >DroEre_CAF1 61177 112 - 1 -------AACGGAAACGGGCCAAGUCUCGACGAGAAUCCAUUUCCAUGUGCUUGGGGUCUGCCUGUGGGAUUGCCUGCAAUUGUGCGGGGAAAAAGAAAACAGAAGCAUAAAAUCAAAU -------...(((((.(((.....((((...)))).))).))))).(((((((..(.(((.((((((.(((((....))))).)))))).....)))...)..)))))))......... ( -32.50) >DroYak_CAF1 62502 119 - 1 AAACGGAAACGGAAACGGGCCAAGUCUCGACGAGAAUCCAUUUCCAUGUGCCUGGGAGCUCCCCGUGGGAUUGCCUGCAAUUGUGCGGGGAUAAAGAAAACAGAAGCAUAAAAUCAAAU ....(((((((....))((.....((((...))))..)).))))).((((((((....(((((((((.(((((....))))).))))))))....)....)))..)))))......... ( -37.30) >consensus _______AACGGAAACGGGCCAAGUCUCGACGAGAAUCCAUUUCCAUGUGCUUUGGA_CUCCCUGUGGGAUUGCCUGCAAUUGUGCGGGGAAAAAGAAAACAGAAGCAUAAAAUCAAAU ..........(((((.(((.....((((...)))).))).))))).((((((((.....((((((((.(((((....))))).))))))))...........))))))))......... (-27.47 = -28.55 + 1.08)

| Location | 20,663,989 – 20,664,085 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.62 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
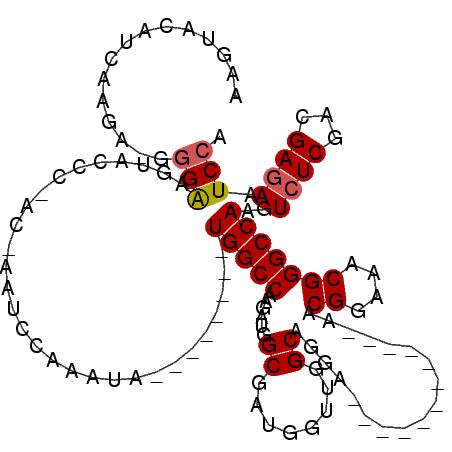
>X_DroMel_CAF1 20663989 96 - 22224390 AAGUACAUCAUGA-GGGAAGUACCC-AC-AAUCCAAAUA--------UGGCCAAGAUGGCGGUGGUUAGCAGGA------------AACGGAAACGGGCCAAGUCUCGACGAAAAUCCA ..((.((((.((.-(((.....)))-.)-).........--------..(((.....))))))).......((.------------..((....))..))........))......... ( -20.60) >DroSec_CAF1 65665 97 - 1 AAGUACAUCAAGAAGAGAAGUACCC-AC-AAUCCAAAUA--------UGGCCAAGAUGGCGAUGGUUGGCAGGA------------AGCGGAAACGGGCCAAGUCUCGACGAGAAUCCA .......((..(..((((....(((-.(-...((((.((--------(.(((.....))).))).))))...).------------...(....)))).....))))..)..))..... ( -24.40) >DroSim_CAF1 43123 97 - 1 AAGUACAUCAAGAAGGGAAGUACCC-UC-AAUCCAAAUA--------UGGCCAAGAUGGCGAUGGUUGGCAGGA------------AGCGGAAACGGGCCAAGUCUCGACGAGAAUCCA ...........((.(((.....)))-))-...((((.((--------(.(((.....))).))).))))..(((------------..((....)).......((((...)))).))). ( -24.80) >DroEre_CAF1 61256 106 - 1 AAGUACAUCAAGA-GGGGAAUACCCCCCAAAAGCAAAUAUGGCGAGAUGGCCAAGAUGGCGAUGGUUGGCAGGA------------AACGGAAACGGGCCAAGUCUCGACGAGAAUCCA ..((.((((....-((((......))))...........((((......)))).)))).(((.(.(((((.(..------------..)(....)..))))).).)))))......... ( -33.40) >DroYak_CAF1 62581 109 - 1 AAGUACUUGAAGA-GGGAAGUAUCC-CCAAAUCCAAGCA--------UGGCCAAGAUGGCUAUGGUUGGCAGGAAACUGAAACGGAAACGGAAACGGGCCAAGUCUCGACGAGAAUCCA .....((((..((-((((....)))-).....((((.((--------(((((.....))))))).))))(((....)))....((...((....))..)).....))..))))...... ( -38.40) >consensus AAGUACAUCAAGA_GGGAAGUACCC_AC_AAUCCAAAUA________UGGCCAAGAUGGCGAUGGUUGGCAGGA____________AACGGAAACGGGCCAAGUCUCGACGAGAAUCCA ...............(((.............................(((((......((........))..................((....)))))))..((((...)))).))). (-17.38 = -17.62 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:55 2006