
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,663,411 – 20,663,549 |
| Length | 138 |
| Max. P | 0.651359 |

| Location | 20,663,411 – 20,663,509 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
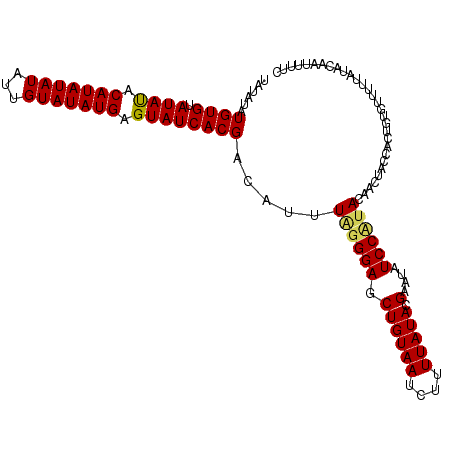
>X_DroMel_CAF1 20663411 98 + 22224390 GAAAAUUU-------------UGGUAGUUGUAAGGAUAUUCGUAUAAAACAUUACAGCUCCCUAAAUGUCGUGAUACUCAUAUACAAUAUAUAUGUAUAUACACAUAUAUA ((..((((-------------.((.(((((((((..(((....)))...).)))))))).)).)))).))((((((..((((((.....))))))..))).)))....... ( -18.40) >DroSec_CAF1 65086 111 + 1 GAAAAUUGUAUAAAAACACAGUGGUAGUUGUAGGGAUAUUCGUAUAAAAGAUUACAGCUCCCCAAAUGUCGUGAUACUCAUAUACAAUAUAUAUGUAUAUACACAUAUAUA ....((((((((...(((((..((.((((((((...(((....))).....)))))))).))....))).))(.....).))))))))((((((((......)))))))). ( -20.50) >DroSim_CAF1 42526 111 + 1 GAAAAUUGUAUAAAAACACAGUGGUAGUUGUAUGGAUAUUCGUAUAAAAGAUUACAGCUCCCAAAAUGUCGUGAUACUCAUAUACAAUAUAUAUGUGUAUACACAUAUAUA ....((((((((...(((((.(((.(((((((....................))))))).)))...))).))(.....).))))))))(((((((((....))))))))). ( -26.05) >consensus GAAAAUUGUAUAAAAACACAGUGGUAGUUGUAAGGAUAUUCGUAUAAAAGAUUACAGCUCCCAAAAUGUCGUGAUACUCAUAUACAAUAUAUAUGUAUAUACACAUAUAUA ......................((.((((((((...(((....))).....)))))))).))...((((.((((((..((((((.....))))))..))).)))))))... (-16.18 = -16.40 + 0.22)


| Location | 20,663,411 – 20,663,509 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.17 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20663411 98 - 22224390 UAUAUAUGUGUAUAUACAUAUAUAUUGUAUAUGAGUAUCACGACAUUUAGGGAGCUGUAAUGUUUUAUACGAAUAUCCUUACAACUACCA-------------AAAUUUUC ((((((((((....))))))))))((((((((..((((...(((((((((....))).))))))..))))..)))....)))))......-------------........ ( -18.30) >DroSec_CAF1 65086 111 - 1 UAUAUAUGUGUAUAUACAUAUAUAUUGUAUAUGAGUAUCACGACAUUUGGGGAGCUGUAAUCUUUUAUACGAAUAUCCCUACAACUACCACUGUGUUUUUAUACAAUUUUC ((((((((((....))))))))))(((((((.((((((.........((((((.((((((....))))).)....))))))...........)))))).)))))))..... ( -20.95) >DroSim_CAF1 42526 111 - 1 UAUAUAUGUGUAUACACAUAUAUAUUGUAUAUGAGUAUCACGACAUUUUGGGAGCUGUAAUCUUUUAUACGAAUAUCCAUACAACUACCACUGUGUUUUUAUACAAUUUUC ((((((((((....))))))))))(((((((((.....)).(((((..(((.((.((((......(((....)))....)))).)).)))..)))))..)))))))..... ( -23.20) >consensus UAUAUAUGUGUAUAUACAUAUAUAUUGUAUAUGAGUAUCACGACAUUUAGGGAGCUGUAAUCUUUUAUACGAAUAUCCAUACAACUACCACUGUGUUUUUAUACAAUUUUC ......((((.((((.(((((((...))))))).)))))))).....((((((.((((((....))))).)....)))))).............................. (-15.94 = -16.17 + 0.23)

| Location | 20,663,431 – 20,663,549 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.65 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -14.30 |
| Energy contribution | -16.46 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20663431 118 - 22224390 AGCAACCCCAAAUAGUGUGGCAUAUUCAAGGCAAAGAUGUUAUAUAUGUGUAUAUACAUAUAUAUUGUA--UAUGAGUAUCACGACAUUUAGGGAGCUGUAAUGUUUUAUACGAAUAUCC (((..(((.((((..((((..(((((((..((((......((((((((((....)))))))))))))).--..)))))))))))..)))).))).)))...((((((.....)))))).. ( -26.20) >DroSec_CAF1 65119 118 - 1 AGCAACCCCAAAUAGUGUGGCAUGUUCAAGGCAAAGAUGGUAUAUAUGUGUAUAUACAUAUAUAUUGUA--UAUGAGUAUCACGACAUUUGGGGAGCUGUAAUCUUUUAUACGAAUAUCC (((..((((((((..(((((..(((.....)))...(..(((((((((((....)))))))))))..).--........)))))..)))))))).)))...................... ( -31.40) >DroSim_CAF1 42559 118 - 1 AGCAACCCCAAAUAGUGUGGCAUGUUCAAGGCAAAGAUGGUAUAUAUGUGUAUACACAUAUAUAUUGUA--UAUGAGUAUCACGACAUUUUGGGAGCUGUAAUCUUUUAUACGAAUAUCC .(((.(((((((...(((((..(((.....)))...(..(((((((((((....)))))))))))..).--........)))))....)))))).).))).................... ( -29.20) >DroEre_CAF1 60722 95 - 1 AGCAACCCCAAAUAGUGUGGCAUAUUCGAAGCAAAGAUGUUCUAUACAGG-----------GUGCUGUUCUAAUGGGUAUCACCACAUUUAAGGAUGUAU----------CCAAAU---- ..............((((((.((((.(........))))).)))))).((-----------((((.(((((((((((.....)).))))..)))))))))----------))....---- ( -22.10) >DroYak_CAF1 62013 95 - 1 AGCAACCCCAAAUAGUGUGGCAUAUUCGAAGCAAAGACCGCAUAUACAGG-----------GUGCUGUACUAAUGGGUAUCACCGCAUUUAAGGAGCCCU----------ACGAAU---- .((...((((..(((((..((((..((........))((.........))-----------))))..))))).)))).....((........)).))...----------......---- ( -21.60) >consensus AGCAACCCCAAAUAGUGUGGCAUAUUCAAGGCAAAGAUGGUAUAUAUGUGUAUA_ACAUAUAUAUUGUA__UAUGAGUAUCACGACAUUUAGGGAGCUGUAAU_UUUUAUACGAAUAUCC .....(((.((((.(((((..(((((((........((((((((((((((....)))))))))))))).....)))))))))).)))))).))).......................... (-14.30 = -16.46 + 2.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:53 2006