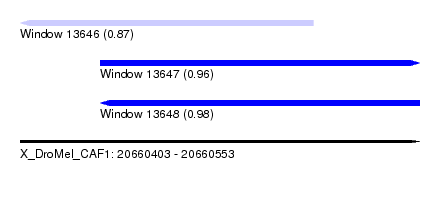
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,660,403 – 20,660,553 |
| Length | 150 |
| Max. P | 0.980992 |

| Location | 20,660,403 – 20,660,513 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20660403 110 - 22224390 AAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAAAUCCACCCCCACCUCC------UCCUACACC--AGACAC--GCCUCC .........(((((.(.((.(((((((((((..........))))))))))).......((.((((...........))))))......------..)).))))--))....--...... ( -28.10) >DroSec_CAF1 62173 116 - 1 AAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAAAUCCACCUCCACCUCCUCCUCCUCCCACACC--AGCCAC--GCCCCC .......(.(((((......(((((((((((..........)))))))))))......(((.(((((.....))))).)))....................)))--)).)..--...... ( -27.90) >DroSim_CAF1 39580 113 - 1 AAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAAAUCCACCUCCACCUCC---UCCUCCCACACC--AGCCAC--GCCCCC .......(.(((((......(((((((((((..........)))))))))))......(((.(((((.....))))).)))........---.........)))--)).)..--...... ( -27.90) >DroEre_CAF1 57691 112 - 1 AAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAUAAGCCACCUCCACCUCC------CC--CUGCCCCUGCCCCUGCCCCCC ...........((..((((.(((((((((((..........))))))))))).....((((.(((......................))------))--))).........))))..)). ( -30.55) >DroYak_CAF1 59001 105 - 1 AAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAA---CUCCUCCACCGCC------CU--CCGCC--AGCCAC--GCCCAU .......(.((((.((.((.(((((((((((..........))))))))))).......(((((.........---........)))))------))--.))))--)).)..--...... ( -31.23) >consensus AAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAAAUCCACCUCCACCUCC______UCC_ACACC__AGCCAC__GCCCCC .......((((.(((...(((((((((((((..........))))))))))).))....))).))))..................................................... (-22.20 = -22.20 + -0.00)



| Location | 20,660,433 – 20,660,553 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -32.16 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20660433 120 + 22224390 GGUGGAUUUUGUUAACCCGCCUGUGGACACACAUCAAUCUCUGUCCGUUGAUUGAUGCGUUCUGCGACCAGUCUUGUUUUUGAAAGCAAAAAUUAAACUCGCACACGAGUAGACGAAAAA (((((.((.....)).)))))((..((.((.((((((((..........)))))))).))))..))....((((.(((((((....)))))))...(((((....)))))))))...... ( -34.10) >DroSec_CAF1 62209 120 + 1 GGUGGAUUUUGUUAACCCGCCUGUGGACACACAUCAAUCUCUGUCCGUUGAUUGAUGCGUUCUGCGACCAGUCUUGUUUUUGAAAGCAAAAAUUAAACUCGCAUACGAGUGGACGAAAAA (((((.((.....)).)))))((..((.((.((((((((..........)))))))).))))..))....((((.(((((((....)))))))...(((((....)))))))))...... ( -34.10) >DroSim_CAF1 39613 120 + 1 GGUGGAUUUUGUUAACCCGCCUGUGGACACACAUCAAUCUCUGUCCGUUGAUUGAUGCGUUCUGCGACCAGUCUUGUUUUUGAAAGCAAAAAUUAAACUCGCAUACGAGUGGACGAAAAA (((((.((.....)).)))))((..((.((.((((((((..........)))))))).))))..))....((((.(((((((....)))))))...(((((....)))))))))...... ( -34.10) >DroEre_CAF1 57723 120 + 1 GGUGGCUUAUGUUAACCCGCCUGUGGACACACAUCAAUCUCUGUCCGUUGAUUGAUGCGUUCUGCGACCAGUCUUGUUUUUGAAAGCAAAAAUUAAACUCGCAUGCGAGUGGACGAAAAA (((((.(((...))).)))))((..((.((.((((((((..........)))))))).))))..))....((((.(((((((....)))))))...(((((....)))))))))...... ( -35.30) >DroYak_CAF1 59029 117 + 1 GGAG---UUUGUUAACCCGCCUGUGGACACACAUCAAUCUCUGUCCGUUGAUUGAUGCGUUCUGCGACCAGUCUUGUUUUUGAAAGCAAAAAUUAAACUCGCAUGUGAGUGGACGAAAGA ((.(---((....)))))...((..((.((.((((((((..........)))))))).))))..))....((((.(((((((....)))))))...(((((....)))))))))...... ( -29.60) >consensus GGUGGAUUUUGUUAACCCGCCUGUGGACACACAUCAAUCUCUGUCCGUUGAUUGAUGCGUUCUGCGACCAGUCUUGUUUUUGAAAGCAAAAAUUAAACUCGCAUACGAGUGGACGAAAAA (((((...........)))))((..((.((.((((((((..........)))))))).))))..))....((((.(((((((....)))))))...(((((....)))))))))...... (-32.16 = -32.24 + 0.08)


| Location | 20,660,433 – 20,660,553 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -31.52 |
| Energy contribution | -31.48 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.980992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20660433 120 - 22224390 UUUUUCGUCUACUCGUGUGCGAGUUUAAUUUUUGCUUUCAAAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAAAUCCACC ......((((..((.(((((.(((((..((((((....))))))..))))).).))))))(((((((((((..........)))))))))))......))))(((((.....)))))... ( -33.70) >DroSec_CAF1 62209 120 - 1 UUUUUCGUCCACUCGUAUGCGAGUUUAAUUUUUGCUUUCAAAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAAAUCCACC ......((..((((((.(((((((((..((((((....))))))..))))...)))))..(((((((((((..........)))))))))))........))))))..)).......... ( -31.70) >DroSim_CAF1 39613 120 - 1 UUUUUCGUCCACUCGUAUGCGAGUUUAAUUUUUGCUUUCAAAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAAAUCCACC ......((..((((((.(((((((((..((((((....))))))..))))...)))))..(((((((((((..........)))))))))))........))))))..)).......... ( -31.70) >DroEre_CAF1 57723 120 - 1 UUUUUCGUCCACUCGCAUGCGAGUUUAAUUUUUGCUUUCAAAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAUAAGCCACC ......((..((((((.(((((((((..((((((....))))))..))))...)))))..(((((((((((..........)))))))))))........))))))..)).......... ( -33.70) >DroYak_CAF1 59029 117 - 1 UCUUUCGUCCACUCACAUGCGAGUUUAAUUUUUGCUUUCAAAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAA---CUCC ......((..((((...(((((((((..((((((....))))))..))))...)))))..(((((((((((..........)))))))))))(((....)))))))..))...---.... ( -28.90) >consensus UUUUUCGUCCACUCGUAUGCGAGUUUAAUUUUUGCUUUCAAAAACAAGACUGGUCGCAGAACGCAUCAAUCAACGGACAGAGAUUGAUGUGUGUCCACAGGCGGGUUAACAAAAUCCACC ......((..((((((.(((((((((..((((((....))))))..))))...)))))..(((((((((((..........)))))))))))........))))))..)).......... (-31.52 = -31.48 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:50 2006