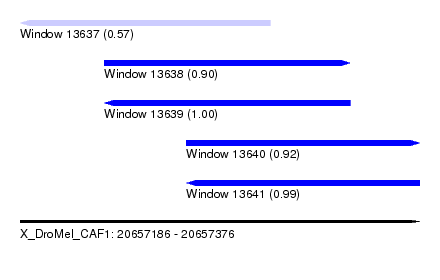
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,657,186 – 20,657,376 |
| Length | 190 |
| Max. P | 0.997313 |

| Location | 20,657,186 – 20,657,305 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -25.78 |
| Energy contribution | -26.09 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20657186 119 - 22224390 GGGGAAGGGUUAAGGGAAAGCCAGGCGUUUUUCCUGAUUACG-GGAAAUACCCUUACCCAUUGAGAGAGAGCUCACAAAUAAUUCAACAAAUCGGAAAGUAUUUUUUCUAAUAUGAGUGG (..(((((((.(((((...((...))...(((((((....))-)))))..))))))))).(((.(((....))).)))....)))..)..((..(((((....)))))..))........ ( -30.20) >DroSec_CAF1 53881 113 - 1 GGGGAAGGGUUAAGGGAAAGCCAGGCGUUUUUCCUGAUUGCG-GAAAAUACCCUUUCCCAUCG------AGCUCUCAAAUAAUUCAACAAAUCGGAAAGUAUUUUUUCUAAUAUGAGUGG ((((((((((..(((((((((.....))))))))).......-......))))))))))....------............(((((....((..(((((....)))))..)).))))).. ( -30.56) >DroEre_CAF1 54631 114 - 1 GGGAAAGGGUUAAGGGAAGGCCAGGCGUUUUUCCUGAUUGCGCGGAAAUACCCUUUCCCAUUGGGC--GGGCUCACCUAUAGUUCAACAAAUGGAGACCAAGU---UUAAAUGUGA-GUG ((((((((((..(((((((((.....)))))))))..............)))))))))).......--..((((((......((.(((....(....)...))---).))..))))-)). ( -33.19) >DroYak_CAF1 55838 114 - 1 GGGGAAGGGUUAAGGGAAAGCCAGGGGUUUUUCCUGAUUGCG-GCAAAUACCCUUUCCCAUUGAGU--GGCCUCACAAACAAUUCAACAAAUUGAGAGGUAUU---UCCAAAAUUAUUUG ((((((((((..((((((((((...)))))))))).......-......))))))))))...(((.--.(((((.(((.............))).)))))..)---))............ ( -35.88) >consensus GGGGAAGGGUUAAGGGAAAGCCAGGCGUUUUUCCUGAUUGCG_GGAAAUACCCUUUCCCAUUGAG___GAGCUCACAAAUAAUUCAACAAAUCGAAAAGUAUU___UCUAAUAUGAGUGG ((((((((((..(((((((((.....)))))))))..............))))))))))...(((......))).............................................. (-25.78 = -26.09 + 0.31)


| Location | 20,657,226 – 20,657,343 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.56 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -30.37 |
| Energy contribution | -30.43 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20657226 117 + 22224390 AUUUGUGAGCUCUCUCUCAAUGGGUAAGGGUAUUUCC-CGUAAUCAGGAAAAACGCCUGGCUUUCCCUUAACCCUUCCCCGGCCCUUGUGGGCCUGUUUUCCCUGUUCGUUUCAGUCU ......((((...........(((.((((((......-...(..((((.......))))..)........)))))).)))(((((....)))))..........)))).......... ( -33.43) >DroSec_CAF1 53921 111 + 1 AUUUGAGAGCU------CGAUGGGAAAGGGUAUUUUC-CGCAAUCAGGAAAAACGCCUGGCUUUCCCUUAACCCUUCCCCGGCCCCUGUGGGCCUGUUUUCCCUGUUCAUUUCAGUCU ....((((((.------....(((.((((((.(((((-(.......))))))......((.....))...)))))).)))(((((....))))).)))))).(((.......)))... ( -35.30) >DroEre_CAF1 54667 116 + 1 AUAGGUGAGCCC--GCCCAAUGGGAAAGGGUAUUUCCGCGCAAUCAGGAAAAACGCCUGGCCUUCCCUUAACCCUUUCCCGGCCCCUUUGGGCCUGUUUUCCCUGUUCAUUUCAGUCU (((((.(((..(--.......((((((((((.(((((.........))))).......((.....))...))))))))))(((((....))))).)..))))))))............ ( -39.20) >DroYak_CAF1 55875 115 + 1 GUUUGUGAGGCC--ACUCAAUGGGAAAGGGUAUUUGC-CGCAAUCAGGAAAAACCCCUGGCUUUCCCUUAACCCUUCCCCGGCCCCUGUGGGCCCGUUUUCCCUGUUCAUUUCAGUCU .....((((...--.))))..(((.((((((......-.....(((((.......)))))..........)))))).)))(((((....)))))........(((.......)))... ( -32.60) >consensus AUUUGUGAGCCC___CUCAAUGGGAAAGGGUAUUUCC_CGCAAUCAGGAAAAACGCCUGGCUUUCCCUUAACCCUUCCCCGGCCCCUGUGGGCCUGUUUUCCCUGUUCAUUUCAGUCU ......((((...........(((.((((((........((...((((.......)))))).........)))))).)))(((((....)))))..........)))).......... (-30.37 = -30.43 + 0.06)


| Location | 20,657,226 – 20,657,343 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 88.56 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -38.95 |
| Energy contribution | -38.89 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20657226 117 - 22224390 AGACUGAAACGAACAGGGAAAACAGGCCCACAAGGGCCGGGGAAGGGUUAAGGGAAAGCCAGGCGUUUUUCCUGAUUACG-GGAAAUACCCUUACCCAUUGAGAGAGAGCUCACAAAU ...(((.......)))........(((((....)))))(((.((((((...((.....)).......(((((((....))-))))).)))))).))).(((.(((....))).))).. ( -38.00) >DroSec_CAF1 53921 111 - 1 AGACUGAAAUGAACAGGGAAAACAGGCCCACAGGGGCCGGGGAAGGGUUAAGGGAAAGCCAGGCGUUUUUCCUGAUUGCG-GAAAAUACCCUUUCCCAUCG------AGCUCUCAAAU ...............((((.....(((((....)))))((((((((((..(((((((((.....))))))))).......-......))))))))))....------...)))).... ( -40.76) >DroEre_CAF1 54667 116 - 1 AGACUGAAAUGAACAGGGAAAACAGGCCCAAAGGGGCCGGGAAAGGGUUAAGGGAAGGCCAGGCGUUUUUCCUGAUUGCGCGGAAAUACCCUUUCCCAUUGGGC--GGGCUCACCUAU ..............(((.......(((((....)))))((((((((((..(((((((((.....)))))))))..............))))))))))..((((.--...))))))).. ( -42.49) >DroYak_CAF1 55875 115 - 1 AGACUGAAAUGAACAGGGAAAACGGGCCCACAGGGGCCGGGGAAGGGUUAAGGGAAAGCCAGGGGUUUUUCCUGAUUGCG-GCAAAUACCCUUUCCCAUUGAGU--GGCCUCACAAAC ...(((.......)))((...((.(((((....)))))((((((((((..((((((((((...)))))))))).......-......)))))))))).....))--..))........ ( -43.56) >consensus AGACUGAAAUGAACAGGGAAAACAGGCCCACAGGGGCCGGGGAAGGGUUAAGGGAAAGCCAGGCGUUUUUCCUGAUUGCG_GGAAAUACCCUUUCCCAUUGAG___GAGCUCACAAAU ...............(((......(((((....)))))((((((((((..(((((((((.....)))))))))..............))))))))))............)))...... (-38.95 = -38.89 + -0.06)



| Location | 20,657,265 – 20,657,376 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -26.75 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20657265 111 + 22224390 UAAUCAGGAAAAACGCCUGGCUUUCCCUUAACCCUUCCCCGGCCCUUGUGGGCCUGUUUUCCCUGUUCGUUUCAGUCUCGGCGUGUUUAACCCUUUUGCCGGGCCACUUUU ....((((.(((((....((.((......)).))......(((((....))))).))))).)))).........(.(((((((.............))))))).)...... ( -29.92) >DroSec_CAF1 53954 111 + 1 CAAUCAGGAAAAACGCCUGGCUUUCCCUUAACCCUUCCCCGGCCCCUGUGGGCCUGUUUUCCCUGUUCAUUUCAGUCUCGACGUGUUUAACCCUUUUGCCGGGCCACUUUU ..............(((((((...................(((((....))))).(((....(((.......)))....)))...............)))))))....... ( -26.60) >DroEre_CAF1 54705 111 + 1 CAAUCAGGAAAAACGCCUGGCCUUCCCUUAACCCUUUCCCGGCCCCUUUGGGCCUGUUUUCCCUGUUCAUUUCAGUCUCGGCGUGUUUAACCCUUUUGCCGGGCCGCUUUU ....((((.(((((....((.....)).............(((((....))))).))))).)))).........(.(((((((.............))))))).)...... ( -28.62) >DroYak_CAF1 55912 110 + 1 CAAUCAGGAAAAACCCCUGGCUUUCCCUUAACCCUUCCCCGGCCCCUGUGGGCCCGUUUUCCCUGUUCAUUUCAGUCUCGGCGUGUUUAACCCUUUUGCCGGGCCACU-UU ....((((.(((((....((.((......)).))......(((((....))))).))))).)))).........(.(((((((.............))))))).)...-.. ( -27.72) >consensus CAAUCAGGAAAAACGCCUGGCUUUCCCUUAACCCUUCCCCGGCCCCUGUGGGCCUGUUUUCCCUGUUCAUUUCAGUCUCGGCGUGUUUAACCCUUUUGCCGGGCCACUUUU ....((((.(((((....((.....)).............(((((....))))).))))).)))).........(.(((((((.............))))))).)...... (-26.75 = -27.00 + 0.25)

| Location | 20,657,265 – 20,657,376 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -35.96 |
| Consensus MFE | -34.61 |
| Energy contribution | -34.68 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20657265 111 - 22224390 AAAAGUGGCCCGGCAAAAGGGUUAAACACGCCGAGACUGAAACGAACAGGGAAAACAGGCCCACAAGGGCCGGGGAAGGGUUAAGGGAAAGCCAGGCGUUUUUCCUGAUUA ....(((((((.......))))....))).................((((((((((.(((((....)))))...(...((((.......))))...))))))))))).... ( -35.70) >DroSec_CAF1 53954 111 - 1 AAAAGUGGCCCGGCAAAAGGGUUAAACACGUCGAGACUGAAAUGAACAGGGAAAACAGGCCCACAGGGGCCGGGGAAGGGUUAAGGGAAAGCCAGGCGUUUUUCCUGAUUG .....((((((.......))))))..((..(((....)))..))..((((((((((.(((((....)))))...(...((((.......))))...))))))))))).... ( -36.60) >DroEre_CAF1 54705 111 - 1 AAAAGCGGCCCGGCAAAAGGGUUAAACACGCCGAGACUGAAAUGAACAGGGAAAACAGGCCCAAAGGGGCCGGGAAAGGGUUAAGGGAAGGCCAGGCGUUUUUCCUGAUUG .....((((.((((...............)))).).))).......((((((((((.(((((....))))).......((((.......))))....)))))))))).... ( -35.56) >DroYak_CAF1 55912 110 - 1 AA-AGUGGCCCGGCAAAAGGGUUAAACACGCCGAGACUGAAAUGAACAGGGAAAACGGGCCCACAGGGGCCGGGGAAGGGUUAAGGGAAAGCCAGGGGUUUUUCCUGAUUG ..-..(((((((((...............)))....(((.......)))......(.(((((....))))).)....))))))((((((((((...))))))))))..... ( -35.96) >consensus AAAAGUGGCCCGGCAAAAGGGUUAAACACGCCGAGACUGAAAUGAACAGGGAAAACAGGCCCACAGGGGCCGGGGAAGGGUUAAGGGAAAGCCAGGCGUUUUUCCUGAUUG .....((((((.......))))))......................((((((((((.(((((....))))).......((((.......))))....)))))))))).... (-34.61 = -34.68 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:40 2006