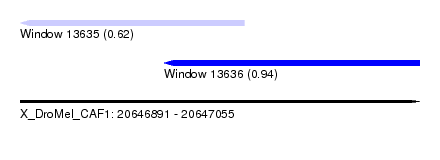
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,646,891 – 20,647,055 |
| Length | 164 |
| Max. P | 0.935790 |

| Location | 20,646,891 – 20,646,983 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 97.61 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20646891 92 - 22224390 AUUCAAUCUGUUUUGCAUAUUGAAUCAAUAAGAGAUUUAAGCCGAGAUGCAACGGGAGAUGACCAGUUUCAAUCCGUCUCUCUCAGUUCCAA .......(((..((((((.(((((((.......)))))))......)))))).((((((((.............)))))))).)))...... ( -19.62) >DroSec_CAF1 51135 92 - 1 AUUCAAUCUGUUUUGCAUAUUGAAUCAAUAAGAGAUUUAAGCCGAGAUGCAACGGGAGAUGACCAGCUUCAAUCCGUCUCUCUCAGUUCCAA .......(((..((((((.(((((((.......)))))))......)))))).((((((((.............)))))))).)))...... ( -19.62) >DroSim_CAF1 33737 92 - 1 AUUCAAUCUGUUUUGCAUAUUGAAUCAAUAAGAGAUUUAAGCCGAGAUGCAACGGGAGAUGACCAGCUUCAAUCCGUCUCUCUCAGUUCCAA .......(((..((((((.(((((((.......)))))))......)))))).((((((((.............)))))))).)))...... ( -19.62) >DroEre_CAF1 51910 92 - 1 AUUCAAUCUGUUUUGCAUAUUGAAUCAAUGAGAGAUUUAAGCCGAGAUGCAACCGGAGAUGACCAGUUUCCAUCCGUCUCUCUCAGUUCCAA (((((((.((.....)).)))))))...(((((((.....((......))....((((((.....)))))).......)))))))....... ( -21.70) >DroYak_CAF1 52962 92 - 1 AUUGAAUCUGUUUUGCAUAUUGAAUCAAUAAGAGAUUUAAGCCGAGAUGCAACGGGAGAUGACCAGUUUCAAUCCGUCUCUCUCAGUUCCAA ...(((.(((..((((((.(((((((.......)))))))......)))))).((((((((.............)))))))).))))))... ( -21.42) >consensus AUUCAAUCUGUUUUGCAUAUUGAAUCAAUAAGAGAUUUAAGCCGAGAUGCAACGGGAGAUGACCAGUUUCAAUCCGUCUCUCUCAGUUCCAA .......(((..((((((.(((((((.......)))))))......)))))).((((((((.............)))))))).)))...... (-18.72 = -18.92 + 0.20)


| Location | 20,646,950 – 20,647,055 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 93.93 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -21.58 |
| Energy contribution | -21.54 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20646950 105 - 22224390 UGAUUUAACGAAAAUCGCUUGUGAA-AAAGGCUUUUAUAAGUGCAGCAAGAGGGUACCGACGAUAUCCUUUGGAUUCAAUCUGUUUUGCAUAUUGAAUCAAUAAGA .(((((.....)))))(((((((((-(.....))))))))))......((((((((.......)))))))).((((((((.((.....)).))))))))....... ( -25.10) >DroSec_CAF1 51194 105 - 1 UGAUUUAACGAAAAUCGCUUGUGAA-AAAGGCUUUUACAAGUGCAGCAAGAGGGGACCGACGAUAUCCUUUGGAUUCAAUCUGUUUUGCAUAUUGAAUCAAUAAGA ((((((((........(((((((((-(.....))))))))))((((..(((..(((((((.(.....).)))).)))..)))...))))...))))))))...... ( -26.90) >DroSim_CAF1 33796 105 - 1 UGAUUUAACGAAAAUCGCUUGUGAA-AAAGGCUUUUACAAGUGCAGCAAGAGGGCACCGACGAUAUCCUUUGGAUUCAAUCUGUUUUGCAUAUUGAAUCAAUAAGA ((((((((.......((((((((((-(.....)))))))))))..((((((.((....((....((((...))))))...)).))))))...))))))))...... ( -25.30) >DroEre_CAF1 51969 105 - 1 UGAUUUAACGAAAAUCGCUUGUGAAUAAAGGCUCUUACAAGUGCAGCA-AGAGGGACCGACGAUAUCCGUUGGAUUCAAUCUGUUUUGCAUAUUGAAUCAAUGAGA ((((((((.......(((((((((..........)))))))))..(((-(((.(((((((((.....))))))......))).))))))...))))))))...... ( -30.40) >DroYak_CAF1 53021 105 - 1 UGAUUUAACGAAAAUCGCUUGUGAA-AAAGACUCUUACAAGUGCAGCAAAGGGGGACCGACGAUAUCGCUUGGAUUGAAUCUGUUUUGCAUAUUGAAUCAAUAAGA ((((((((((.....))..((..((-(.(((.((...((((((.......((....))........))))))....)).))).)))..))..))))))))...... ( -25.96) >consensus UGAUUUAACGAAAAUCGCUUGUGAA_AAAGGCUUUUACAAGUGCAGCAAGAGGGGACCGACGAUAUCCUUUGGAUUCAAUCUGUUUUGCAUAUUGAAUCAAUAAGA ((((((((.......((((((((((........))))))))))..((((((.((......(((......)))........)).))))))...))))))))...... (-21.58 = -21.54 + -0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:36 2006