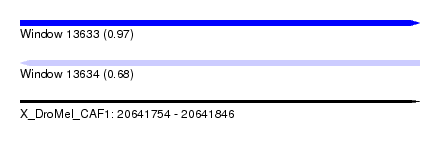
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,641,754 – 20,641,846 |
| Length | 92 |
| Max. P | 0.974179 |

| Location | 20,641,754 – 20,641,846 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -21.61 |
| Energy contribution | -21.43 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20641754 92 + 22224390 UCUCGGAAUAUGUCCGGGUGCAAAUUAUUAAAGAUUGUGUGGAAAAUAAGUUUUUCAUGUUUCCCAAAAACUUUAUCCGCUGACAGUCCGAA ..(((((...((((((((((..................((((((((....))))))))((((.....))))..))))))..)))).))))). ( -22.30) >DroSec_CAF1 46017 92 + 1 UCUCGGAAUGUGUCCGGGUGCAAAUUAUUAUAGAUUGUGUGGAAAAUAAGUUUUUCAUGUUCACCAAAAACUUUAUCCGCUGACAGUCCAAA .((((((.....))))))..............(((((((((((....((((((((..........))))))))..)))))..)))))).... ( -21.80) >DroEre_CAF1 46798 87 + 1 UCCCGGAAUGUGUCCGGGUGCAAAUUAUUAAAGAUUGUGUGGGAAAUAAGUUUUUCAUGUUUCCCAAAAACUUUAUCCGCUGCCAGU----- .((((((.....)))))).(((......(((((.((...(((((((((.(.....).))))))))).)).))))).....)))....----- ( -25.80) >DroYak_CAF1 47664 92 + 1 UCCCGGAAUGUGUCCGGGUGCAAAUUAUUAAAGAUUGUGUGGAAAAUAAGUUUUUCAUUUUUCCCAAAAACUUUAUCCGCUGCCAGUCAGUA .((((((.....))))))..............(((((.(((((....((((((((..........))))))))..)))))...))))).... ( -24.80) >consensus UCCCGGAAUGUGUCCGGGUGCAAAUUAUUAAAGAUUGUGUGGAAAAUAAGUUUUUCAUGUUUCCCAAAAACUUUAUCCGCUGACAGUCCGAA .((((((.....))))))..............(((((.(((((....((((((((..........))))))))..)))))...))))).... (-21.61 = -21.43 + -0.19)



| Location | 20,641,754 – 20,641,846 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -15.79 |
| Energy contribution | -16.47 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20641754 92 - 22224390 UUCGGACUGUCAGCGGAUAAAGUUUUUGGGAAACAUGAAAAACUUAUUUUCCACACAAUCUUUAAUAAUUUGCACCCGGACAUAUUCCGAGA ((((((.((((.((((((((((.((.(((((((.((((.....))))))))).)).)).)))))....))))).....))))...)))))). ( -21.00) >DroSec_CAF1 46017 92 - 1 UUUGGACUGUCAGCGGAUAAAGUUUUUGGUGAACAUGAAAAACUUAUUUUCCACACAAUCUAUAAUAAUUUGCACCCGGACACAUUCCGAGA ((((((.((((.(.(((..((((((((.((....)).))))))))....))).).(((...........)))......))))...)))))). ( -17.90) >DroEre_CAF1 46798 87 - 1 -----ACUGGCAGCGGAUAAAGUUUUUGGGAAACAUGAAAAACUUAUUUCCCACACAAUCUUUAAUAAUUUGCACCCGGACACAUUCCGGGA -----.......((((((((((....((((((..((((.....))))))))))......)))))....))))).((((((.....)))))). ( -23.00) >DroYak_CAF1 47664 92 - 1 UACUGACUGGCAGCGGAUAAAGUUUUUGGGAAAAAUGAAAAACUUAUUUUCCACACAAUCUUUAAUAAUUUGCACCCGGACACAUUCCGGGA ............((((((((((.((.((((((((.(((.....))))))))).)).)).)))))....))))).((((((.....)))))). ( -22.70) >consensus UUCGGACUGGCAGCGGAUAAAGUUUUUGGGAAACAUGAAAAACUUAUUUUCCACACAAUCUUUAAUAAUUUGCACCCGGACACAUUCCGAGA ............((((((((((....((((((..((((.....))))))))))......)))))....))))).((((((.....)))))). (-15.79 = -16.47 + 0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:34 2006