| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,603,359 – 20,603,495 |
| Length | 136 |
| Max. P | 0.796238 |

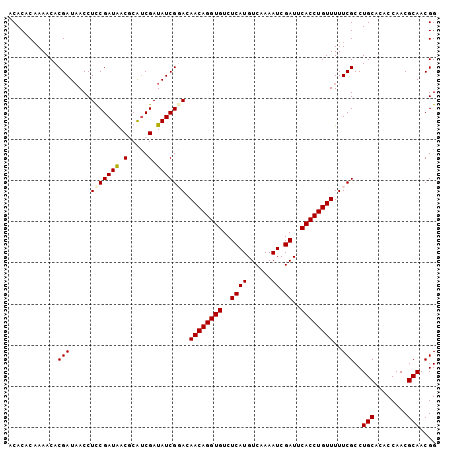
| Location | 20,603,359 – 20,603,463 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20603359 104 - 22224390 GCACACUAAACACGAUAACCUCCGAUGACUCAACGAUAUCGUAUAACAGGUGUCUCAUGUCAAAAUCGAUUCACCUGUUUUUCGCCUGCACACCAACGCAACGG ((.........((((((.(....((....))...).))))))..((((((((..((((......)).))..))))))))....)).(((........))).... ( -18.30) >DroSec_CAF1 8044 104 - 1 GCACACAAAACACGAUAACCUCCGAUGACUCAUCGAUAUCGUAUAACAGGUGUCUCAUGUCAAAAUCGAUUCACCUGUUUUUCGCCUGCACACCAACGCAACGG ((.........((((((.....(((((...))))).))))))..((((((((..((((......)).))..))))))))....)).(((........))).... ( -22.60) >DroEre_CAF1 8074 104 - 1 ACACACAAAACACGAUAACCUCCGAUAACGCGUCGAUAUCGGACAACAGGUGUCUCAUGUCAAAAUCGAUUCACCUGUUUUUCGCCUGCACACCAACGCAACGG ............(((.....(((((((.((...)).))))))).((((((((..((((......)).))..))))))))..)))..(((........))).... ( -23.60) >DroYak_CAF1 8290 104 - 1 ACACACAAAACACGAUUACCUUCGAUAACGCAUCGAUAUCGGACAACAGGUGUCUCAUGUCAAAAUCGAUUCACCUGUUUUUCGCCUGCACACCAACGCAACGG ............((.......(((((.....)))))...((((.((((((((..((((......)).))..)))))))).))))..(((........))).)). ( -21.80) >consensus ACACACAAAACACGAUAACCUCCGAUAACGCAUCGAUAUCGGACAACAGGUGUCUCAUGUCAAAAUCGAUUCACCUGUUUUUCGCCUGCACACCAACGCAACGG ............(((.....(((((((.(.....).))))))).((((((((..((((......)).))..))))))))..)))..(((........))).... (-19.09 = -19.27 + 0.19)


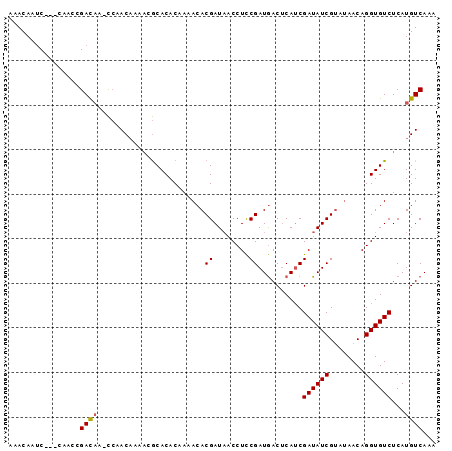
| Location | 20,603,399 – 20,603,495 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -13.28 |
| Consensus MFE | -9.05 |
| Energy contribution | -9.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20603399 96 - 22224390 AAACAACCAACCAACCGACAAUCCAACAAAACGCACACUAAACACGAUAACCUCCGAUGACUCAACGAUAUCGUAUAACAGGUGUCUCAUGUCAAA ................((((......................((((........)).)).......((((((........))))))...))))... ( -10.90) >DroSec_CAF1 8084 92 - 1 AAACAAUC----AACCGACAACCCAACAAAACGCACACAAAACACGAUAACCUCCGAUGACUCAUCGAUAUCGUAUAACAGGUGUCUCAUGUCAAA ........----....((((............((((.......((((((.....(((((...))))).)))))).......))))....))))... ( -12.63) >DroYak_CAF1 8330 89 - 1 AAACAAUCUGUCGGUUGAU---UGAAC-C---ACACACAAAACACGAUUACCUUCGAUAACGCAUCGAUAUCGGACAACAGGUGUCUCAUGUCAAA ...(((((........)))---))...-.---............(((......)))..........(((((.(((((.....))))).)))))... ( -16.30) >consensus AAACAAUC___CAACCGACAA_CCAACAAAACGCACACAAAACACGAUAACCUCCGAUGACUCAUCGAUAUCGUAUAACAGGUGUCUCAUGUCAAA ................((((........................((........))..........((((((........))))))...))))... ( -9.05 = -9.17 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:06 2006