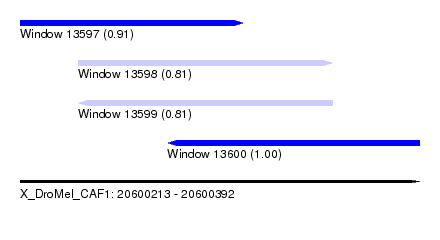
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,600,213 – 20,600,392 |
| Length | 179 |
| Max. P | 0.997801 |

| Location | 20,600,213 – 20,600,313 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.18 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20600213 100 + 22224390 GGACG--------------AGCGCAGCUGGGAGUGGUGGGCAUAAAAACUGUCUGACAUAUCCGCUGGUCGAUUUGAUUAAGUUGUCCCAUC------GCGACCAGCUGCCACGGCUUAU ....(--------------(((((((((((..((((((((.((((.....(((((((..........))))....)))....))))))))))------))..))))))))....)))).. ( -39.40) >DroPse_CAF1 5679 113 + 1 GGUCGGCCGUUGGGGCAUUGUCACAGCU-GGAGUUAUGGAAAUAAAAACUGUCUGCCAUAUCCGUUUGCCGAUUUGAUUAAGUUGUCAGAGG------AGGCUCCCGGAGGACCCAAGAG (((((((.((((.(((...))).)))).-(...((((....))))...).....)))...((((...(((..((((((......))))))..------.)))...)))).))))...... ( -33.10) >DroSec_CAF1 4755 99 + 1 GGUCG--------------AGCGCAGCA-GGAGUGGUGGGCAUAAAAACUGUCUGACAUAUCCGCUGGUCGACUUGAUUAAGUUGUCCCAUC------GCGACCAGCUGCCUUGGCUUAC (((((--------------((.(((((.-((.((((((((((........(((.(((..........))))))..........)).))))))------))..)).))))))))))))... ( -38.47) >DroEre_CAF1 4688 105 + 1 GGACG--------------AGCGCAGCU-GGAGUGGUGGGCAUAAAAACUGUCUGACAUAUCCGCUGGUCGAUUUGAUUAAGUUGUCCCAUCGACCAUGCGACCAGCUGCCUUGGCACAC ...((--------------((.((((((-((.(((..(((((.......)))))..)))...(((((((((((..(((......)))..)))))))).))).))))))))))))...... ( -46.30) >DroYak_CAF1 4827 99 + 1 GGACG--------------AGCGCAGCU-GGAGUGGUGGACAUAAAAACUGUCUGACAUAUCCGCUGGUCGAUUUGAUUAAGUUGUCCCAUC------GCGACCAGCUGCCUUGGCACAC ...((--------------((.((((((-((.((((((((((.......)))).............((.((((((....)))))).))))))------))..))))))))))))...... ( -36.60) >consensus GGACG______________AGCGCAGCU_GGAGUGGUGGGCAUAAAAACUGUCUGACAUAUCCGCUGGUCGAUUUGAUUAAGUUGUCCCAUC______GCGACCAGCUGCCUUGGCACAC .....................((((((..((((((..(((((.......)))))..))).))))))((.((((((....)))))).))..........)))....(((.....))).... (-18.70 = -18.18 + -0.52)

| Location | 20,600,239 – 20,600,353 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.21 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -24.34 |
| Energy contribution | -27.96 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20600239 114 + 22224390 CAUAAAAACUGUCUGACAUAUCCGCUGGUCGAUUUGAUUAAGUUGUCCCAUC------GCGACCAGCUGCCACGGCUUAUAGCUGGCCAAAAGAAUAGGACAAAUUUCGGGCUUGGUUUU ....((((((((((((.((.(((((((((((....(((......))).....------.)))))))).((((..((.....))))))..........)))...)).))))))..)))))) ( -33.70) >DroSec_CAF1 4780 114 + 1 CAUAAAAACUGUCUGACAUAUCCGCUGGUCGACUUGAUUAAGUUGUCCCAUC------GCGACCAGCUGCCUUGGCUUACAGCUGGCCAAGAUAAUAGGACAAAUUUCGGAUUUGGUUUU ....((((((((((((.((.(((((((((((....(((......))).....------.))))))))...(((((((.......)))))))......)))...)).))))))..)))))) ( -37.40) >DroEre_CAF1 4713 114 + 1 CAUAAAAACUGUCUGACAUAUCCGCUGGUCGAUUUGAUUAAGUUGUCCCAUCGACCAUGCGACCAGCUGCCUUGGCACACAGCGAGCCAAAAUAUUAGGAGCAAUGU------GGGUUUU ............((.((((...(((((((((((..(((......)))..)))))))).)))....(((.(((((((.(.....).)))).......)))))).))))------.)).... ( -33.11) >DroYak_CAF1 4852 108 + 1 CAUAAAAACUGUCUGACAUAUCCGCUGGUCGAUUUGAUUAAGUUGUCCCAUC------GCGACCAGCUGCCUUGGCACACAGCUGGCCAAAAUAAUAGGACAAAUGU------GCGUUUU ....(((((.((...((((.(((((((((((....(((......))).....------.))))))))....(((((.((....))))))).......)))...))))------))))))) ( -28.40) >consensus CAUAAAAACUGUCUGACAUAUCCGCUGGUCGAUUUGAUUAAGUUGUCCCAUC______GCGACCAGCUGCCUUGGCACACAGCUGGCCAAAAUAAUAGGACAAAUGU______GGGUUUU ....(((((((((((((((.(((((((((((....(((......)))............))))))))....(((((.........))))).......)))...)))))))))..)))))) (-24.34 = -27.96 + 3.62)

| Location | 20,600,239 – 20,600,353 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.21 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -26.04 |
| Energy contribution | -27.10 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20600239 114 - 22224390 AAAACCAAGCCCGAAAUUUGUCCUAUUCUUUUGGCCAGCUAUAAGCCGUGGCAGCUGGUCGC------GAUGGGACAACUUAAUCAAAUCGACCAGCGGAUAUGUCAGACAGUUUUUAUG ..........(((....((((((((((....(((((((((....((....))))))))))).------)))))))))).....((.....))....)))..................... ( -29.30) >DroSec_CAF1 4780 114 - 1 AAAACCAAAUCCGAAAUUUGUCCUAUUAUCUUGGCCAGCUGUAAGCCAAGGCAGCUGGUCGC------GAUGGGACAACUUAAUCAAGUCGACCAGCGGAUAUGUCAGACAGUUUUUAUG .....((.(((((.....(((((((((....(((((((((((........))))))))))).------)))))))))((((....)))).......))))).))................ ( -36.30) >DroEre_CAF1 4713 114 - 1 AAAACCC------ACAUUGCUCCUAAUAUUUUGGCUCGCUGUGUGCCAAGGCAGCUGGUCGCAUGGUCGAUGGGACAACUUAAUCAAAUCGACCAGCGGAUAUGUCAGACAGUUUUUAUG .......------((((...(((.........((((.(((((.(....).))))).))))((.((((((((..((........))..))))))))))))).))))............... ( -34.00) >DroYak_CAF1 4852 108 - 1 AAAACGC------ACAUUUGUCCUAUUAUUUUGGCCAGCUGUGUGCCAAGGCAGCUGGUCGC------GAUGGGACAACUUAAUCAAAUCGACCAGCGGAUAUGUCAGACAGUUUUUAUG ....(((------....((((((((((....(((((((((((.(....).))))))))))).------)))))))))).....((.....))...)))...................... ( -34.60) >consensus AAAACCA______AAAUUUGUCCUAUUAUUUUGGCCAGCUGUAAGCCAAGGCAGCUGGUCGC______GAUGGGACAACUUAAUCAAAUCGACCAGCGGAUAUGUCAGACAGUUUUUAUG (((((............((((((((((....(((((((((((........))))))))))).......))))))))))............(((..........))).....))))).... (-26.04 = -27.10 + 1.06)


| Location | 20,600,279 – 20,600,392 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -24.01 |
| Energy contribution | -25.07 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20600279 113 - 22224390 AAGAGGCACUUUUAUAAGAAUUAAAAUGAA-UAAAUACAAAAAACCAAGCCCGAAAUUUGUCCUAUUCUUUUGGCCAGCUAUAAGCCGUGGCAGCUGGUCGC------GAUGGGACAACU ..(.(((.(((....)))........((..-......)).........)))).....((((((((((....(((((((((....((....))))))))))).------)))))))))).. ( -30.30) >DroSec_CAF1 4820 112 - 1 AAGA-GCACUUUUAUAAGAAUUAAAAUGUA-UGAAUACAAAAAACCAAAUCCGAAAUUUGUCCUAUUAUCUUGGCCAGCUGUAAGCCAAGGCAGCUGGUCGC------GAUGGGACAACU ....-(((.(((((.......)))))))).-..........................((((((((((....(((((((((((........))))))))))).------)))))))))).. ( -31.50) >DroEre_CAF1 4753 114 - 1 AAGAAACGUUUCUAAGAUAAUUAAAAUAAAUUGAAUACAAAAAACCC------ACAUUGCUCCUAAUAUUUUGGCUCGCUGUGUGCCAAGGCAGCUGGUCGCAUGGUCGAUGGGACAACU .((((....))))................................((------.(((((..((........(((((.(((((.(....).))))).)))))...)).)))))))...... ( -20.30) >DroYak_CAF1 4892 108 - 1 AAGAAAACAUUUUAAAAGAAUUAAAAUGAAGUGAAUACAAAAAACGC------ACAUUUGUCCUAUUAUUUUGGCCAGCUGUGUGCCAAGGCAGCUGGUCGC------GAUGGGACAACU .......((((((((.....))))))))..(((.............)------))..((((((((((....(((((((((((.(....).))))))))))).------)))))))))).. ( -36.22) >consensus AAGAAACACUUUUAAAAGAAUUAAAAUGAA_UGAAUACAAAAAACCA______AAAUUUGUCCUAUUAUUUUGGCCAGCUGUAAGCCAAGGCAGCUGGUCGC______GAUGGGACAACU .........................................................((((((((((....(((((((((((........))))))))))).......)))))))))).. (-24.01 = -25.07 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:04 2006