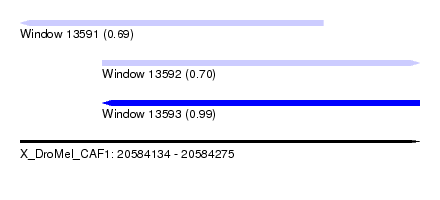
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,584,134 – 20,584,275 |
| Length | 141 |
| Max. P | 0.986031 |

| Location | 20,584,134 – 20,584,241 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.76 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -24.56 |
| Energy contribution | -24.53 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20584134 107 - 22224390 --CUUUGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUAACAUUUGGCCUGAAAGUGAUAGAUUAUAUGGAGACCGAUCAGGAAGAUCGCAUGG----------- --((((.(((((((((((((....((..(((((....))))).....))....))))))))))))))))).............(....)(((((.....))))).....----------- ( -30.00) >DroVir_CAF1 203278 109 - 1 UGUUGCAUAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCUUGGAUGUGAUAGAUUAUAUGGAGACCGAUCAGGAAGAUCGCAUGG----------- (((..(((.(((((((((((....((..(((((....))))).....))....)))))))))))...)))..)))........(....)(((((.....))))).....----------- ( -28.70) >DroPse_CAF1 188028 106 - 1 ---UUUGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCCUGGAUGUGAUAGAUUAUAUGGAGACCGAUCAAGAAGACCGCAUGG----------- ---..(((.((((....(.(((....))).)((((((((.(((...(((((((.((((((.....)))))).))))))).))).))))..)))).....)))))))...----------- ( -27.50) >DroGri_CAF1 183205 109 - 1 CGUUGCAUAGGUCAAAUGUUUCUUAUGAAUAAAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCUUGGAUGUGAUAGAUUAUAUGGAGACCGAUCAGGAAGAUCGCAUGG----------- ...(((...((((((((..(((....)))...)))).((.(((...(((((((.((((((.....)))))).))))))).))).))))))((((.....)))))))...----------- ( -27.80) >DroSim_CAF1 168319 118 - 1 --CUUUGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCCUGAAAGUGAUAGAUUAUAUGGAGACCGAUCAGGAAGAUCGCAUGGAUCCGNNNNNN --((((.(((((((((((((....((..(((((....))))).....))....))))))))))))))))).............(((..((((((.....)))))...)..)))....... ( -34.40) >DroWil_CAF1 189176 108 - 1 -CAAACGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCUUAGAUGUGAUAGAUUAUAUGGAGACCGAUCAGGAAGAUCGUAUGG----------- -((.((...((((............((.(((((....)))))..))(((((((.(((((((...))))))).))))))).......))))((((.....)))))).)).----------- ( -26.20) >consensus __CUUCGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCCUGGAUGUGAUAGAUUAUAUGGAGACCGAUCAGGAAGAUCGCAUGG___________ .......(((((((((((((....((..(((((....))))).....))....)))))))))))))...(((...........(....)(((((.....))))))))............. (-24.56 = -24.53 + -0.03)


| Location | 20,584,163 – 20,584,275 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.98 |
| Mean single sequence MFE | -14.40 |
| Consensus MFE | -8.10 |
| Energy contribution | -8.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20584163 112 + 22224390 UAAUCUAUCACUUUCAGGCCAAAUGUUAUAAAUUCGCCAUCGAAAAAUCGAUUCAUAAGAAACAUUUGACCUGCAAAG---GAACCACAAAAAAGAU--ACAAAUCAGCACCAAAGA- ...(((.((.((((((((.((((((((...........(((((....)))))((....)))))))))).)))).))))---))...........(((--....)))........)))- ( -20.40) >DroVir_CAF1 203307 101 + 1 UAAUCUAUCACAUCCAAGCCAAAUGUCAUAAAUUCGCCAUCGAAAAAUCGAUUCAUAAGAAACAUUUGACCUAUGCAACAAGAACGACAUAUA-------C--A-------CACAGA- .................((.....((((....(((...(((((....)))))......))).....))))....)).................-------.--.-------......- ( -8.30) >DroPse_CAF1 188057 106 + 1 UAAUCUAUCACAUCCAGGCCAAAUGUCAUAAAUUCGCCAUCGAAAAAUCGAUUCAUAAGAAACAUUUGACCUGCAAA----GAAACACAAACAGAAU--UUAGAUC-----CAAUUC- ..............((((.(((((((......(((...(((((....)))))......)))))))))).))))....----................--.......-----......- ( -14.50) >DroWil_CAF1 189205 102 + 1 UAAUCUAUCACAUCUAAGCCAAAUGUCAUAAAUUCGCCAUCGAAAAAUCGAUUCAUAAGAAACAUUUGACCUGCGUUUG--AAAGAACAAAGAAGAC--------------CAGAGAA ...(((......(((..(((((((((......(((...(((((....)))))......))))))))))....))((((.--...))))..)))....--------------...))). ( -13.92) >DroMoj_CAF1 252474 94 + 1 UAAUCUAUCACAUCCAAGCCAAAUGUCAUAAAUUCACCAUCGAAAAAUCGAUUCAUAAGAAACAUUUGACCUGCGAAACGAAAACAACAAAUU-----------------------A- ............((((.(.(((((((.............((((....))))(((....)))))))))).).)).)).................-----------------------.- ( -9.80) >DroAna_CAF1 160954 117 + 1 UAAUCUAUCACAUCCAGGCCAAAUGUCAUAAAUUCGCCAUCGAAAAAUCGAUUCAUAAGAAACAUUUGACCUGAAAAAGGUGAAACAAAAACAAAAUGUAGCAAUUAGCAAGUCAGC- .......((((...((((.(((((((......(((...(((((....)))))......)))))))))).))))......)))).................((.....))........- ( -19.50) >consensus UAAUCUAUCACAUCCAAGCCAAAUGUCAUAAAUUCGCCAUCGAAAAAUCGAUUCAUAAGAAACAUUUGACCUGCAAAA___GAACCACAAACAA_AU______A_______CAAAGA_ ...................(((((((.............((((....))))(((....)))))))))).................................................. ( -8.10 = -8.10 + -0.00)



| Location | 20,584,163 – 20,584,275 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.98 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -21.15 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20584163 112 - 22224390 -UCUUUGGUGCUGAUUUGU--AUCUUUUUUGUGGUUC---CUUUGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUAACAUUUGGCCUGAAAGUGAUAGAUUA -.....(((((......))--))).........((..---((((.(((((((((((((....((..(((((....))))).....))....)))))))))))))))))..))...... ( -31.00) >DroVir_CAF1 203307 101 - 1 -UCUGUG-------U--G-------UAUAUGUCGUUCUUGUUGCAUAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCUUGGAUGUGAUAGAUUA -......-------.--.-------............((((..(((.(((((((((((....((..(((((....))))).....))....)))))))))))...)))..)))).... ( -23.90) >DroPse_CAF1 188057 106 - 1 -GAAUUG-----GAUCUAA--AUUCUGUUUGUGUUUC----UUUGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCCUGGAUGUGAUAGAUUA -(((((.-----......)--))))....(.((((..----..(.(((((((((((((....((..(((((....))))).....))....))))))))))))).)...)))).)... ( -26.50) >DroWil_CAF1 189205 102 - 1 UUCUCUG--------------GUCUUCUUUGUUCUUU--CAAACGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCUUAGAUGUGAUAGAUUA .......--------------...........(((.(--((.....((((((((((((....((..(((((....))))).....))....)))))))))))).....))).)))... ( -22.00) >DroMoj_CAF1 252474 94 - 1 -U-----------------------AAUUUGUUGUUUUCGUUUCGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGUGAAUUUAUGACAUUUGGCUUGGAUGUGAUAGAUUA -(-----------------------(((((((..(.......((.(((((((((((((..((((..(((((....))))).))))......))))))))))))))).)..)))))))) ( -25.80) >DroAna_CAF1 160954 117 - 1 -GCUGACUUGCUAAUUGCUACAUUUUGUUUUUGUUUCACCUUUUUCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCCUGGAUGUGAUAGAUUA -((......))(((..((........))..)))..((((....(((((((((((((((....((..(((((....))))).....))....))))))))))))))).))))....... ( -29.70) >consensus _UCUGUG_______U______AU_UUAUUUGUGGUUC___UUUCGCAGGUCAAAUGUUUCUUAUGAAUCGAUUUUUCGAUGGCGAAUUUAUGACAUUUGGCCUGGAUGUGAUAGAUUA .............................................(((((((((((((....((..(((((....))))).....))....))))))))))))).............. (-21.15 = -20.82 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:59 2006