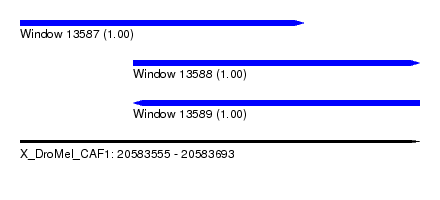
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,583,555 – 20,583,693 |
| Length | 138 |
| Max. P | 0.999894 |

| Location | 20,583,555 – 20,583,653 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.20 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.70 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20583555 98 + 22224390 GU-GUCACGGAAAAAUCUAAGGCUUAAAUUUCACAUAGGGCUCUACGAAGUAUGUAGUGAAAAAAAAA---------------------UUGUUUUGGCUACAUACUUCUAGGCACCUAC ..-(((..(((....)))..)))............((((((.(((.((((((((((((.((((.....---------------------...)))).))))))))))))))))).)))). ( -31.50) >DroSec_CAF1 153353 115 + 1 GU-GUCACAGAAAAAUCUAAGGCUUCAAUUUCACCUAGGGGUCUACGAAGUAUGCAGAAUAAAACGAACACGAAUGCGAAAUAUAUAUUUUGUUUUGGCUGCAUACUUCUAGGCAC---- ((-(((.............(((((((...........)))))))..(((((((((((..(((((((((....................))))))))).)))))))))))..)))))---- ( -32.35) >DroSim_CAF1 167327 115 + 1 GU-GUCACAGAAAAAUCUAAGGCUUCAAUUUCACCUAGGGGUCUACGAAGUAUGCAGUGAAAAACGAACUCGAAUGCGAAAUAUAUAUUUUGUUUUGGCUGCAUACUUCUAGGCAC---- ((-(((.............(((((((...........)))))))..((((((((((((.(((..((....))...(((((((....)))))))))).))))))))))))..)))))---- ( -36.30) >DroEre_CAF1 147543 114 + 1 GUGGCACCAGAAAGAGCUAAGGUUUCAAAUUCACAUAGGGGUCUACAAGGUAUGCAGUGAAAAACGAACACGAAUGCGAACUAGAUA--GUAUUUUUGCUGCAUACCACUAGACAC---- .(((.(((............))).))).............(((((...(((((((((..((((.......((....)).(((....)--)).))))..)))))))))..)))))..---- ( -29.00) >DroYak_CAF1 150515 112 + 1 GU-GCCACAGCAAGAGCUAAGGUUUCAAAUUCGCAUAG-GGUCUACAAAGUAUGCAGUGAAAGACGAACUCGAGUGCGAACUAGUAU--AUGUUUAUGCUGCAUACUUCUAGACAC---- ..-(((..(((....)))..)))...............-.(((((..(((((((((((...(((((......(((....))).....--.)))))..))))))))))).)))))..---- ( -29.70) >consensus GU_GUCACAGAAAAAUCUAAGGCUUCAAUUUCACAUAGGGGUCUACGAAGUAUGCAGUGAAAAACGAACACGAAUGCGAAAUAGAUA__UUGUUUUGGCUGCAUACUUCUAGGCAC____ ...(((..(((....)))..))).................(((((.((((((((((((.((((((((......................)))))))))))))))))))))))))...... (-22.37 = -22.93 + 0.56)



| Location | 20,583,594 – 20,583,693 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -26.28 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.20 |
| Structure conservation index | 0.80 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20583594 99 + 22224390 CUCUACGAAGUAUGUAGUGAAAAAAAAA---------------------UUGUUUUGGCUACAUACUUCUAGGCACCUACCUAUACGAGUAUAGGGAUUUUUGUGGAACCCACAUACAUA .((((.((((((((((((.((((.....---------------------...)))).))))))))))))))))......((((((....))))))......(((((...)))))...... ( -32.40) >DroSec_CAF1 153392 111 + 1 GUCUACGAAGUAUGCAGAAUAAAACGAACACGAAUGCGAAAUAUAUAUUUUGUUUUGGCUGCAUACUUCUAGGCAC----CUAUACGAGUAUAUUGAUU-UUGUGGAACCCACA----UA (((((.(((((((((((..(((((((((....................))))))))).))))))))))))))))..----...................-.(((((...)))))----.. ( -31.35) >DroSim_CAF1 167366 112 + 1 GUCUACGAAGUAUGCAGUGAAAAACGAACUCGAAUGCGAAAUAUAUAUUUUGUUUUGGCUGCAUACUUCUAGGCAC----CUAUACGAGUAUAUUGAUUUUUGUGGAACCCACA----UA (((((.((((((((((((.(((..((....))...(((((((....)))))))))).)))))))))))))))))..----.....................(((((...)))))----.. ( -35.30) >consensus GUCUACGAAGUAUGCAGUGAAAAACGAAC_CGAAUGCGAAAUAUAUAUUUUGUUUUGGCUGCAUACUUCUAGGCAC____CUAUACGAGUAUAUUGAUUUUUGUGGAACCCACA____UA (((((.(((((((((((...((((((((....................))))))))..))))))))))))))))...........................(((((...)))))...... (-26.28 = -26.62 + 0.34)

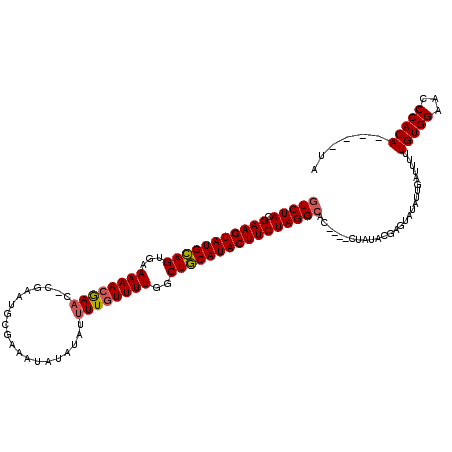

| Location | 20,583,594 – 20,583,693 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -22.08 |
| Energy contribution | -21.97 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20583594 99 - 22224390 UAUGUAUGUGGGUUCCACAAAAAUCCCUAUACUCGUAUAGGUAGGUGCCUAGAAGUAUGUAGCCAAAACAA---------------------UUUUUUUUUCACUACAUACUUCGUAGAG ...((((((((...)))).......((((((....))))))...))))((((((((((((((..((((...---------------------.....))))..))))))))))).))).. ( -27.40) >DroSec_CAF1 153392 111 - 1 UA----UGUGGGUUCCACAA-AAUCAAUAUACUCGUAUAG----GUGCCUAGAAGUAUGCAGCCAAAACAAAAUAUAUAUUUCGCAUUCGUGUUCGUUUUAUUCUGCAUACUUCGUAGAC ..----(((((...))))).-........(((.....(((----....)))(((((((((((..(((((.((((....))))(((....)))...)))))...))))))))))))))... ( -26.00) >DroSim_CAF1 167366 112 - 1 UA----UGUGGGUUCCACAAAAAUCAAUAUACUCGUAUAG----GUGCCUAGAAGUAUGCAGCCAAAACAAAAUAUAUAUUUCGCAUUCGAGUUCGUUUUUCACUGCAUACUUCGUAGAC ..----(((((...)))))..........(((.....(((----....)))(((((((((((..(((((...........((((....))))...)))))...))))))))))))))... ( -24.84) >consensus UA____UGUGGGUUCCACAAAAAUCAAUAUACUCGUAUAG____GUGCCUAGAAGUAUGCAGCCAAAACAAAAUAUAUAUUUCGCAUUCG_GUUCGUUUUUCACUGCAUACUUCGUAGAC ......(((((...)))))...............((((......))))((((((((((((((..(((((..........................)))))...))))))))))).))).. (-22.08 = -21.97 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:55 2006