
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,537,521 – 20,537,648 |
| Length | 127 |
| Max. P | 0.999916 |

| Location | 20,537,521 – 20,537,612 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 95.71 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20537521 91 + 22224390 CAUCAAAUGGCAAACACGCUGGCGUUUUAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCUCGGCAGCAGCAAAAAACUGACACAUCAGCA .................((((..((.(((((.(((......)))(((((((((((........)))))))))))..))))).))..)))). ( -28.60) >DroSec_CAF1 105029 91 + 1 CAUCAAAUGGCAAACACGCUGUCGUUUUAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCUCGGCAGCAGCAAAAAACUGACACAGCAGCA .................(((((.((.(((((.(((......)))(((((((((((........)))))))))))..))))).)).))))). ( -29.40) >DroSim_CAF1 114395 91 + 1 CAUCAAAUGGCAAACACGCUGGCGUUUUAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCUCGGCAGCAGCAAAAAACUGACACAGCAGCA .................((((..((.(((((.(((......)))(((((((((((........)))))))))))..))))).))..)))). ( -26.60) >DroEre_CAF1 103917 91 + 1 CAUCAAAUGGCAAACACGCUUGCAUUUCAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCGCGGCAGCAGCAAAAAACUGACACUGCAGCA .................(((.(((..(((((.(((......)))(((((((((((........)))))))))))..)))))...)))))). ( -27.70) >DroYak_CAF1 106876 91 + 1 CAUCAAAUGGCAAACACGCUCGCGUUCUAGUAUUGAUUAUGCAGUUUGUUGCUGCGAAUCUCGGCAGCAGCAAAAAACUGACACAGCAGCA .................(((.((......((((.....))))..(((((((((((........)))))))))))...........))))). ( -24.80) >consensus CAUCAAAUGGCAAACACGCUGGCGUUUUAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCUCGGCAGCAGCAAAAAACUGACACAGCAGCA .................((((..((.(((((.(((......)))(((((((((((........)))))))))))..))))).))..)))). (-24.92 = -25.56 + 0.64)

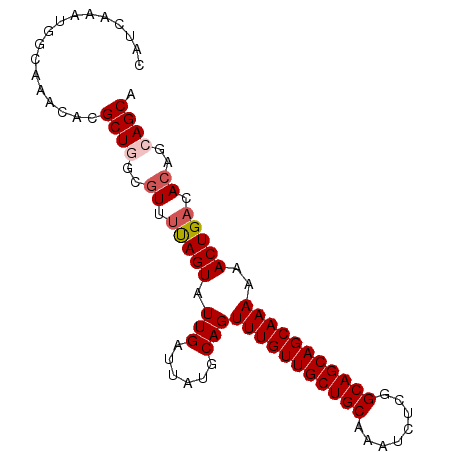

| Location | 20,537,521 – 20,537,612 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 95.71 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -26.98 |
| Energy contribution | -27.06 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.54 |
| SVM RNA-class probability | 0.999916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20537521 91 - 22224390 UGCUGAUGUGUCAGUUUUUUGCUGCUGCCGAGAUUUGCAGCAACAAACUGCAUAAUCAAUACUAAAACGCCAGCGUGUUUGCCAUUUGAUG ......((((.((((((.(((((((...........))))))).))))))))))(((((.....((((((....)))))).....))))). ( -28.70) >DroSec_CAF1 105029 91 - 1 UGCUGCUGUGUCAGUUUUUUGCUGCUGCCGAGAUUUGCAGCAACAAACUGCAUAAUCAAUACUAAAACGACAGCGUGUUUGCCAUUUGAUG ......((((.((((((.(((((((...........))))))).))))))))))(((((.....(((((......))))).....))))). ( -25.60) >DroSim_CAF1 114395 91 - 1 UGCUGCUGUGUCAGUUUUUUGCUGCUGCCGAGAUUUGCAGCAACAAACUGCAUAAUCAAUACUAAAACGCCAGCGUGUUUGCCAUUUGAUG ......((((.((((((.(((((((...........))))))).))))))))))(((((.....((((((....)))))).....))))). ( -28.50) >DroEre_CAF1 103917 91 - 1 UGCUGCAGUGUCAGUUUUUUGCUGCUGCCGCGAUUUGCAGCAACAAACUGCAUAAUCAAUACUGAAAUGCAAGCGUGUUUGCCAUUUGAUG .(((((((((.((((((.(((((((...........))))))).)))))))))..(((....)))..))).)))................. ( -26.50) >DroYak_CAF1 106876 91 - 1 UGCUGCUGUGUCAGUUUUUUGCUGCUGCCGAGAUUCGCAGCAACAAACUGCAUAAUCAAUACUAGAACGCGAGCGUGUUUGCCAUUUGAUG ......((((.((((((.(((((((...........))))))).))))))))))(((((.....((((((....)))))).....))))). ( -29.40) >consensus UGCUGCUGUGUCAGUUUUUUGCUGCUGCCGAGAUUUGCAGCAACAAACUGCAUAAUCAAUACUAAAACGCCAGCGUGUUUGCCAUUUGAUG ......((((.((((((.(((((((...........))))))).))))))))))(((((.....((((((....)))))).....))))). (-26.98 = -27.06 + 0.08)

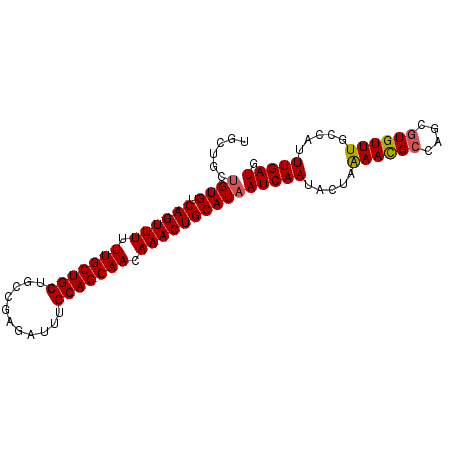

| Location | 20,537,548 – 20,537,648 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20537548 100 + 22224390 UAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCUCGGCAGCAGCAAAAAACUGACACAUCAGCAGCACCAACUGCACAACAGCAACACACGCCGAGAAAA ............((((((.....))))))..(((((((.((((.......))).)......((((......))))...............)))))))... ( -28.10) >DroSec_CAF1 105056 100 + 1 UAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCUCGGCAGCAGCAAAAAACUGACACAGCAGCAGCACCAACUACACAUCAGCCACACACGCCGAGAAAA ............((((((.....))))))..(((((((.((((.......))).)...((....))........................)))))))... ( -24.10) >DroSim_CAF1 114422 100 + 1 UAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCUCGGCAGCAGCAAAAAACUGACACAGCAGCAGCACCAACUGCACAUCAGCCACACACGCCGAGAAAA ............((((((.....))))))..(((((((.((((.......))).)......((((......))))...............)))))))... ( -28.10) >DroEre_CAF1 103944 99 + 1 CAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCGCGGCAGCAGCAAAAAACUGACACUGCAGCAGCACCAACUGCACUUCAGCAGCACACGUCGCGAAA- ......(((...(((((((((((((((((....((....)).(((......)).)..))))))))))..)))))))..)))((.((....)).))....- ( -30.50) >DroYak_CAF1 106903 99 + 1 UAGUAUUGAUUAUGCAGUUUGUUGCUGCGAAUCUCGGCAGCAGCAAAAAACUGACACAGCAGCAACACCAACUGCACAUCAGCCACACACGGCGAGAAA- ......((((..((((((((((((((((.....((((.............))))....)))))))))..))))))).))))(((......)))......- ( -31.82) >consensus UAGUAUUGAUUAUGCAGUUUGUUGCUGCAAAUCUCGGCAGCAGCAAAAAACUGACACAGCAGCAGCACCAACUGCACAUCAGCCACACACGCCGAGAAAA ............((((((.....))))))..(((((((.((((.......))).)......((((......))))...............)))))))... (-24.60 = -24.60 + -0.00)


| Location | 20,537,548 – 20,537,648 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -28.18 |
| Energy contribution | -28.46 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20537548 100 - 22224390 UUUUCUCGGCGUGUGUUGCUGUUGUGCAGUUGGUGCUGCUGAUGUGUCAGUUUUUUGCUGCUGCCGAGAUUUGCAGCAACAAACUGCAUAAUCAAUACUA ............((((((..(((((((((((..((((((.(((.((.((((........)))).))..))).))))))...))))))))))))))))).. ( -36.50) >DroSec_CAF1 105056 100 - 1 UUUUCUCGGCGUGUGUGGCUGAUGUGUAGUUGGUGCUGCUGCUGUGUCAGUUUUUUGCUGCUGCCGAGAUUUGCAGCAACAAACUGCAUAAUCAAUACUA ...((((((((.(((.((((((..((((((.......))))).)..))))))...)))...))))))))..(((((.......)))))............ ( -32.00) >DroSim_CAF1 114422 100 - 1 UUUUCUCGGCGUGUGUGGCUGAUGUGCAGUUGGUGCUGCUGCUGUGUCAGUUUUUUGCUGCUGCCGAGAUUUGCAGCAACAAACUGCAUAAUCAAUACUA ...((((((((.(((.((((((..((((((.......))))).)..))))))...)))...))))))))..(((((.......)))))............ ( -34.00) >DroEre_CAF1 103944 99 - 1 -UUUCGCGACGUGUGCUGCUGAAGUGCAGUUGGUGCUGCUGCAGUGUCAGUUUUUUGCUGCUGCCGCGAUUUGCAGCAACAAACUGCAUAAUCAAUACUG -.((((((........))).)))((((((((..((((((....(((.((((........)))).))).....))))))...))))))))........... ( -32.70) >DroYak_CAF1 106903 99 - 1 -UUUCUCGCCGUGUGUGGCUGAUGUGCAGUUGGUGUUGCUGCUGUGUCAGUUUUUUGCUGCUGCCGAGAUUCGCAGCAACAAACUGCAUAAUCAAUACUA -......((((....))))((((((((((((..(((((((((.(((.((((.....)))).))).(.....)))))))))))))))))).))))...... ( -35.50) >consensus UUUUCUCGGCGUGUGUGGCUGAUGUGCAGUUGGUGCUGCUGCUGUGUCAGUUUUUUGCUGCUGCCGAGAUUUGCAGCAACAAACUGCAUAAUCAAUACUA ............((((...((((((((((((..((((((.....((.((((........)))).))......))))))...)))))))).)))))))).. (-28.18 = -28.46 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:39 2006