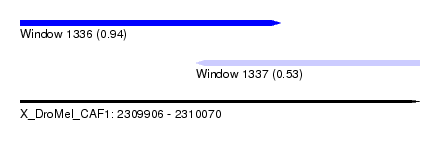
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,309,906 – 2,310,070 |
| Length | 164 |
| Max. P | 0.943040 |

| Location | 2,309,906 – 2,310,013 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -24.26 |
| Energy contribution | -23.94 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
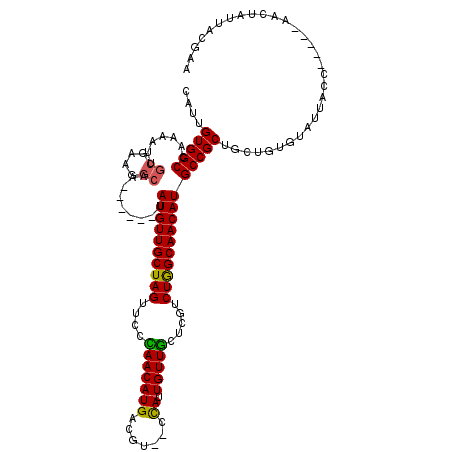
>X_DroMel_CAF1 2309906 107 + 22224390 CAUUGUGGCCAAAAUUGCGAUGGCAA------UAUGUUGCUAGUUCCCAACAUGACGU--CCAUUGUUGCUCGUCUGGCAACAUGCCGCUUCUGUGUAUUACC-----AACUAUUACGAA (((.(((((....(((((....))))------)((((((((((....(((((((....--.)).))))).....)))))))))))))))....))).......-----............ ( -30.10) >DroSec_CAF1 7883 107 + 1 CAUUGUGGCCAAAAUUGCGCAGGCAA------UAUGUUGCUAGUUCCCAACAUGACGU--CCAUUGUUGCUCGUCUGGCAACAUGCCGCCGCUGUGUAUUACC-----AACUAUUACGAA (((.(((((....(((((....))))------)((((((((((....(((((((....--.)).))))).....))))))))))...))))).))).......-----............ ( -31.90) >DroSim_CAF1 8688 107 + 1 CAUUGUGGCCAAAAUUGCGCAGGCAA------UAUGUUGCUAGUUCCCAACAUGACGU--CCAUUGUUGCUCGUCUGGCAACAUGCCGCCGCUGUGUAUUACC-----AACUAUUACGAG (((.(((((....(((((....))))------)((((((((((....(((((((....--.)).))))).....))))))))))...))))).))).......-----............ ( -31.90) >DroEre_CAF1 9141 113 + 1 UUUUGUGGCUAAAAUGGCUUAGGCAUUUUACUUAUGUUGCUAGUUUCUAACAUGACGU--CUAUUGUUGUUCGUCUGGCAACAUGCCGCUGCCGUGUAUUACC-----AACUAUUACCCA .....(((.(((.(((((...((((.........(((((((((...(.((((..(((.--....))))))).).)))))))))))))...)))))...)))))-----)........... ( -29.80) >DroYak_CAF1 8791 116 + 1 UAUUGUGGCUUAAAU----AAGGCAUUUUACUUAUGUUGCUAGUUCCUAACAUGUUGUUUCCAUUGUUAUUUGGCUAGCAACAUGCCGCUGCUGUGUGUUACCAUUACAACUAUUACCCA ....(((((....((----(((........)))))((((((((((..(((((...((....)).)))))...))))))))))..))))).....((((.......))))........... ( -28.50) >consensus CAUUGUGGCCAAAAUUGCGAAGGCAA______UAUGUUGCUAGUUCCCAACAUGACGU__CCAUUGUUGCUCGUCUGGCAACAUGCCGCUGCUGUGUAUUACC_____AACUAUUACGAA ....(((((.......((....)).........((((((((((....(((((((.......)).))))).....)))))))))))))))............................... (-24.26 = -23.94 + -0.32)

| Location | 2,309,978 – 2,310,070 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -19.18 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
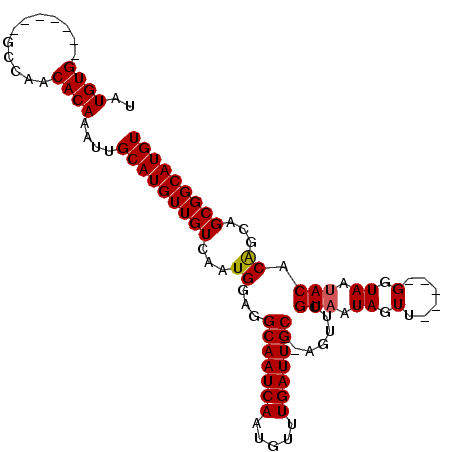
>X_DroMel_CAF1 2309978 92 - 22224390 UAUGUG-------GCCAACACAAAUUGCAUGUUGUCAAUGGAGGCAAUCAAUGUUUGAUUGC--AGUUCGUAAUAGUU-----GGUAAUACACAGAAGCGGCAUGU ..((((-------.....))))....(((((((((..(((((.(((((((.....)))))))--..))))).....((-----(........)))..))))))))) ( -24.70) >DroSec_CAF1 7955 94 - 1 UAUGUG-------GUCAGCACAAAUUGCAUGUUGUCAAUGGAGGCAAUCAAUGUUUGAUUGCACAGUUCGUAAUAGUU-----GGUAAUACACAGCGGCGGCAUGU ..((((-------.....))))....((((((((((.(((((.(((((((.....)))))))....)))))....(((-----(........)))))))))))))) ( -29.90) >DroSim_CAF1 8760 94 - 1 UAUGUG-------GCCAACACAAAUUGCAUGUUGUCAAUGGAGGCAAUCAAUGUUUGAUUGCACAGCUCGUAAUAGUU-----GGUAAUACACAGCGGCGGCAUGU ..((((-------.....))))....((((((((((....((((((((((.....)))))))....)))......(((-----(........)))))))))))))) ( -30.30) >DroEre_CAF1 9219 92 - 1 UCUGUG-------GCCAACACAAAUUGCAUGUUGUCAAUGGAGGCAAUCAAUGUUUGAUUGC--AGUGGGUAAUAGUU-----GGUAAUACACGGCAGCGGCAUGU ..((((-------.....))))...(((.(((((((.......(((((((.....)))))))--.(((..........-----.......)))))))))))))... ( -25.63) >DroYak_CAF1 8867 104 - 1 UCUGUGGCUGGUGGCCAACACAAAUUGCAUGUUGUCAAUGGAGGCAAUCAAUGUUUGAUUGC--AGUGGGUAAUAGUUGUAAUGGUAACACACAGCAGCGGCAUGU .((((.(((((((((((..((((......((((..(.((....(((((((.....)))))))--.)).)..)))).))))..))))..))).)))).))))..... ( -30.60) >consensus UAUGUG_______GCCAACACAAAUUGCAUGUUGUCAAUGGAGGCAAUCAAUGUUUGAUUGC__AGUUCGUAAUAGUU_____GGUAAUACACAGCAGCGGCAUGU ..((((............))))....(((((((((...((...(((((((.....))))))).......(((.((.(......).)).))).))...))))))))) (-19.18 = -19.22 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:44 2006