
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,527,484 – 20,527,579 |
| Length | 95 |
| Max. P | 0.897972 |

| Location | 20,527,484 – 20,527,579 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 88.20 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.05 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
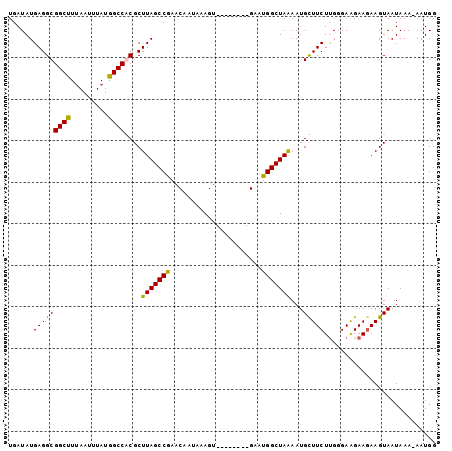
>X_DroMel_CAF1 20527484 95 + 22224390 UGAUAUGAGGCGGCUUUAAUUUAUGGCCGCGCUUAGCCGAACAAUAAAGU--------GAAUGGCUAAAAUGCUUCUCGGGAAGAAGAAGUAAUAAAAAAUGG .........((((((.........))))))..(((((((.((......))--------...)))))))..((((((((.....).)))))))........... ( -25.60) >DroSec_CAF1 95116 95 + 1 UGAUAUGAGGCGGCUUUAAUUUAUGGCCGCGCUUAGCCGAACAAUAAAGU--------GAAUGGCUAAAAUGCUUCUUGGGAAGAAGAAGUAAUAAAAAAUGG .........((((((.........))))))..(((((((.((......))--------...)))))))..((((((((......))))))))........... ( -26.60) >DroEre_CAF1 94091 102 + 1 UGAUAUGAGGCGGCUUAAAUUUAUGGCCACGCUUAGCCGAACAAUAAAGUUGGCUAAAGACUGGCUAAAAUGUUUCUUGUGAGGAAGAAGUAAUAAA-AAUGG ............((((.......((((((..(((((((.(((......))))))).)))..)))))).....(((((.....)))))))))......-..... ( -21.50) >DroYak_CAF1 96788 93 + 1 UGAUAUGAGGCGGCCUUAAUUUAUGGCCACGCUUAGCCAAACAAUAAAGC--------GACUGGCUAGAAUGUUUCUUGGGAAGAA-AAGUAAUAAA-AGUGG ......(((((((((.........))))....(((((((..(........--------)..)))))))...)))))..........-..........-..... ( -19.00) >consensus UGAUAUGAGGCGGCUUUAAUUUAUGGCCACGCUUAGCCGAACAAUAAAGU________GAAUGGCUAAAAUGCUUCUUGGGAAGAAGAAGUAAUAAA_AAUGG ......(((((((((.........))))....(((((((......................)))))))...)))))........................... (-17.86 = -17.05 + -0.81)


| Location | 20,527,484 – 20,527,579 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.20 |
| Mean single sequence MFE | -19.15 |
| Consensus MFE | -13.75 |
| Energy contribution | -14.31 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
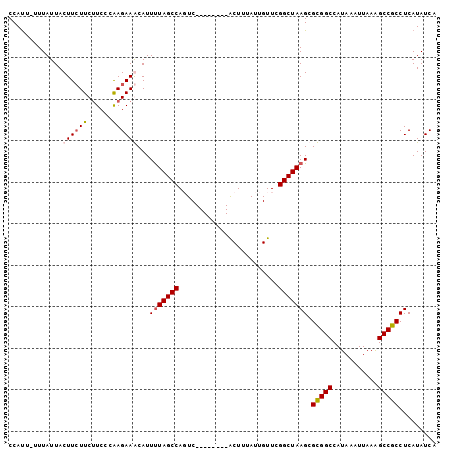
>X_DroMel_CAF1 20527484 95 - 22224390 CCAUUUUUUAUUACUUCUUCUUCCCGAGAAGCAUUUUAGCCAUUC--------ACUUUAUUGUUCGGCUAAGCGCGGCCAUAAAUUAAAGCCGCCUCAUAUCA .............((((((......))))))...(((((((...(--------(......))...))))))).(((((...........)))))......... ( -19.80) >DroSec_CAF1 95116 95 - 1 CCAUUUUUUAUUACUUCUUCUUCCCAAGAAGCAUUUUAGCCAUUC--------ACUUUAUUGUUCGGCUAAGCGCGGCCAUAAAUUAAAGCCGCCUCAUAUCA .............((((((......))))))...(((((((...(--------(......))...))))))).(((((...........)))))......... ( -19.40) >DroEre_CAF1 94091 102 - 1 CCAUU-UUUAUUACUUCUUCCUCACAAGAAACAUUUUAGCCAGUCUUUAGCCAACUUUAUUGUUCGGCUAAGCGUGGCCAUAAAUUUAAGCCGCCUCAUAUCA .....-........(((((......)))))........((((((..(((((((((......))).)))))))).))))......................... ( -17.20) >DroYak_CAF1 96788 93 - 1 CCACU-UUUAUUACUU-UUCUUCCCAAGAAACAUUCUAGCCAGUC--------GCUUUAUUGUUUGGCUAAGCGUGGCCAUAAAUUAAGGCCGCCUCAUAUCA .....-.........(-((((.....))))).....(((((((..--------((......)))))))))((.((((((.........))))))))....... ( -20.20) >consensus CCAUU_UUUAUUACUUCUUCUUCCCAAGAAACAUUUUAGCCAGUC________ACUUUAUUGUUCGGCUAAGCGCGGCCAUAAAUUAAAGCCGCCUCAUAUCA .............((((((......))))))...(((((((........................))))))).(((((...........)))))......... (-13.75 = -14.31 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:34 2006