
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,509,119 – 20,509,231 |
| Length | 112 |
| Max. P | 0.936641 |

| Location | 20,509,119 – 20,509,216 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -27.54 |
| Energy contribution | -27.94 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20509119 97 - 22224390 CCUGCGACCUUGACCAAUCGCUGGCAAGGUCGGGCGGCAGCG-UACGACCUUGAACGGCCCACACAAACCAAAUGGACCAAGGUGUUGACCAGCAAAA .(.((.((((((.......((((.((((((((..((....))-..))))))))..)))).........((....))..)))))))).).......... ( -31.40) >DroSec_CAF1 77232 97 - 1 CCUGCGACCUUGACCAAUCGCAGGCAAGGUCGGGCGGCAGCG-GACGACCUUGAAAGGGCCACACUAACCAAAUGGACCAAGGUGUUGACCAGCAAAA (((((((..........)))))))((((((((..((....))-..))))))))...((.(.(((((..((....)).....))))).).))....... ( -32.90) >DroSim_CAF1 101087 97 - 1 CCUGCGACCUUGACCAAUCGCAGGCAAGGUCGGGCGGCAGCG-GACGACCUUGAAAGGCCCACACUAACCAAAUGGACCAAGGUGUUGACCAGCAAAA (((((((..........)))))))((((((((..((....))-..))))))))...((.(.(((((..((....)).....))))).).))....... ( -32.90) >DroEre_CAF1 76026 97 - 1 CCUGCGACCUUGACCAAUCGAAGGCAAGGUCGGGCGGCAGAGGGACGACCUUGA-CGGCCCACACAAACCAAAUGGACCAAGGUGUUGACCAGCAAAA ..((((((((((.((.......)))))))))((((.(..((((.....))))..-).)))).......((....)).....(((....))).)))... ( -35.60) >DroYak_CAF1 78282 97 - 1 CCUGCGACCUUGACCAAUCGAAGGCAAGGUCGGGCGGCAGCGGGACGACCUUGA-CGGCCCACACAAACCAAAUGAACCAAGGUGUUGACCAGCGAAA ..((((((((((.((.......)))))))))((((.((((.((.....))))).-).))))....................(((....))).)))... ( -32.20) >consensus CCUGCGACCUUGACCAAUCGCAGGCAAGGUCGGGCGGCAGCG_GACGACCUUGAACGGCCCACACAAACCAAAUGGACCAAGGUGUUGACCAGCAAAA ....((((((((.((.......)))))))))).((((((((......((((((...(.....).....((....))..)))))))))).)).)).... (-27.54 = -27.94 + 0.40)

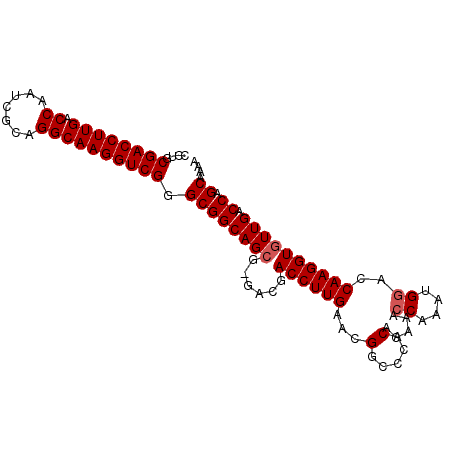
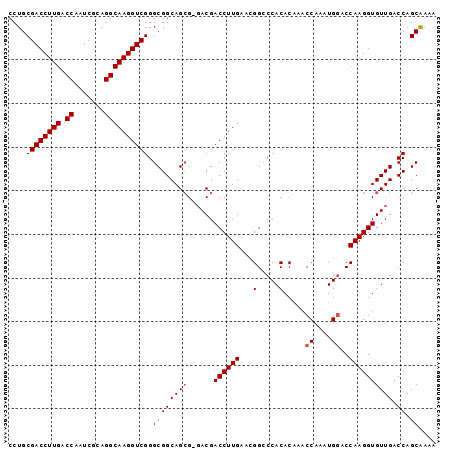
| Location | 20,509,139 – 20,509,231 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 94.60 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -25.46 |
| Energy contribution | -25.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20509139 92 - 22224390 UCACCAAAUCGAGGACCUGCGACCUUGACCAAUCGCUGGCAAGGUCGGGCGGCAGCG-UACGACCUUGAACGGCCCACACAAACCAAAUGGAC ...(((..((((((........))))))......((((.((((((((..((....))-..))))))))..))))..............))).. ( -31.10) >DroSec_CAF1 77252 92 - 1 UCACCAAAUCGAGGACCUGCGACCUUGACCAAUCGCAGGCAAGGUCGGGCGGCAGCG-GACGACCUUGAAAGGGCCACACUAACCAAAUGGAC ...(((.........(((((((..........)))))))((((((((..((....))-..))))))))...((..........))...))).. ( -29.70) >DroSim_CAF1 101107 92 - 1 UCACCAAAUCGAGGACCUGCGACCUUGACCAAUCGCAGGCAAGGUCGGGCGGCAGCG-GACGACCUUGAAAGGCCCACACUAACCAAAUGGAC ............((.(((((((..........)))))))((((((((..((....))-..))))))))......)).......((....)).. ( -29.10) >DroEre_CAF1 76046 92 - 1 UCACCAAAUCGAGGACCUGCGACCUUGACCAAUCGAAGGCAAGGUCGGGCGGCAGAGGGACGACCUUGA-CGGCCCACACAAACCAAAUGGAC ...(((..............(((((((.((.......)))))))))((((.(..((((.....))))..-).))))............))).. ( -32.60) >DroYak_CAF1 78302 92 - 1 UCACCAAAUCGAGGACCUGCGACCUUGACCAAUCGAAGGCAAGGUCGGGCGGCAGCGGGACGACCUUGA-CGGCCCACACAAACCAAAUGAAC ............((......(((((((.((.......)))))))))((((.((((.((.....))))).-).)))).......))........ ( -29.80) >consensus UCACCAAAUCGAGGACCUGCGACCUUGACCAAUCGCAGGCAAGGUCGGGCGGCAGCG_GACGACCUUGAACGGCCCACACAAACCAAAUGGAC ...(((..((((((.....((((((((.((.......)))))))))).((....)).......))))))..((..........))...))).. (-25.46 = -25.86 + 0.40)

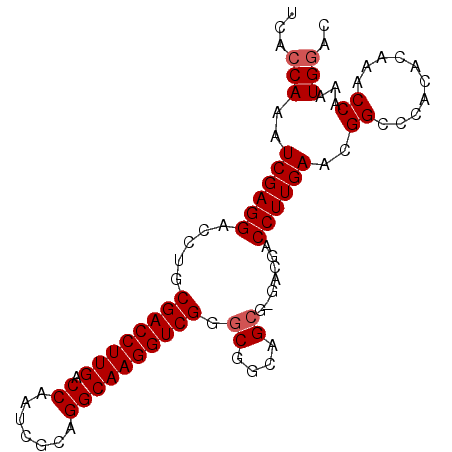
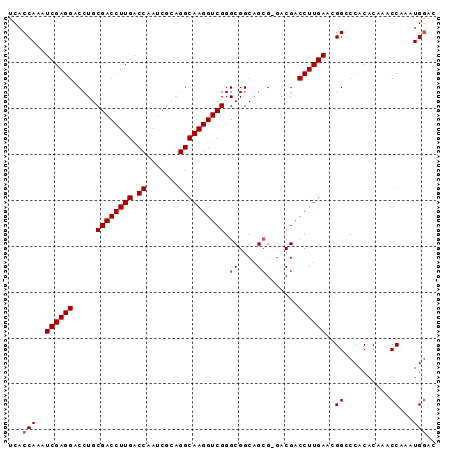
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:30 2006