
| Sequence ID | X_DroMel_CAF1 |
|---|---|
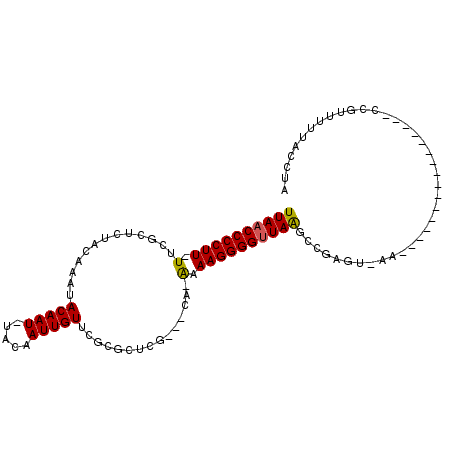
| Location | 20,463,445 – 20,463,546 |
| Length | 101 |
| Max. P | 0.991924 |

| Location | 20,463,445 – 20,463,546 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.17 |
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -12.55 |
| Energy contribution | -12.36 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20463445 101 + 22224390 UUAACCCCUUUUUUGCUCUACAAAUACAAU-UACAAUUGUUCGCGCUCG---CG-AAAAGGGGUUAAGGCGGGUAAAUUUCAAAUUUCACUGACCGUUUUUACCUA (((((((((((..............(((((-....)))))((((....)---))-))))))))))))...(((((((..(((........))).....))))))). ( -26.80) >DroPse_CAF1 58054 84 + 1 UUAACCCCUU-UUCGCUCUACAAAUACAAUUUGCAAUUGUUC-CGCCCGGUCCA-GAAAGGGGUUAAGCUGAGUGAA----------------CCG---UUACCUA ((((((((((-((.((................))...((..(-(....))..))-)))))))))))).....((((.----------------...---))))... ( -19.59) >DroSec_CAF1 36826 76 + 1 UUAACCCCUU-UCUGCUCUACAAAUACAAU-UACAAUUGUUCGCGCUCG---CG-AAAAGGGGCUAAGGCGA------------------------UUUUUACCUA ....((((((-(.............(((((-....)))))((((....)---))-))))))))...(((...------------------------......))). ( -16.50) >DroSim_CAF1 41927 100 + 1 UUAACCCCUU-UCUGCUCUACAAAUACAAU-UACAAUUGUUCGCGCUCG---CG-AAAAGGGGUUAAGGCGGGUAAAUUUCGAAUUUCACUGACCGUUUUUACCUA ((((((((((-(.............(((((-....)))))((((....)---))-))))))))))))...(((((((...((............))..))))))). ( -27.40) >DroAna_CAF1 16077 83 + 1 UUAACCCCUU-UUCGCUCUACAAAUACAAUUUGCAAUUGUUAGCGUGGG---CACAAAAGGGGUUAGGCCUG-------------------GGCCAUUCGUACCUA .(((((((((-((.((.((((....(((((.....)))))....)))))---)..)))))))))))(((...-------------------.)))........... ( -29.40) >DroPer_CAF1 58260 84 + 1 UUAACCCCUU-UUCGCUCUACAAAUACAAUUUGCAAUUGUUC-CGCCCGGUCCA-GAAAGGGGUUAAGCUGAGUGAA----------------CCG---UUACCUA ((((((((((-((.((................))...((..(-(....))..))-)))))))))))).....((((.----------------...---))))... ( -19.59) >consensus UUAACCCCUU_UUCGCUCUACAAAUACAAU_UACAAUUGUUCGCGCUCG___CA_AAAAGGGGUUAAGCCGAGU_AA________________CCGUUUUUACCUA ((((((((((.(.............(((((.....)))))...............).))))))))))....................................... (-12.55 = -12.36 + -0.19)


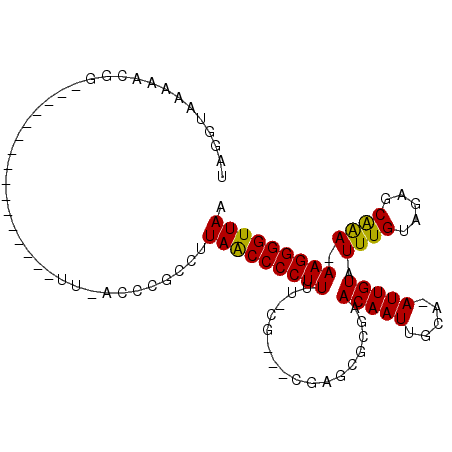
| Location | 20,463,445 – 20,463,546 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.17 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.50 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20463445 101 - 22224390 UAGGUAAAAACGGUCAGUGAAAUUUGAAAUUUACCCGCCUUAACCCCUUUU-CG---CGAGCGCGAACAAUUGUA-AUUGUAUUUGUAGAGCAAAAAAGGGGUUAA ..((((((.....((((......))))..)))))).....(((((((((((-((---(....))))(((((....-))))).((((.....)))))))))))))). ( -27.70) >DroPse_CAF1 58054 84 - 1 UAGGUAA---CGG----------------UUCACUCAGCUUAACCCCUUUC-UGGACCGGGCG-GAACAAUUGCAAAUUGUAUUUGUAGAGCGAA-AAGGGGUUAA .......---...----------------...........((((((((((.-((..((....)-).((((.(((.....))).))))....)).)-))))))))). ( -20.80) >DroSec_CAF1 36826 76 - 1 UAGGUAAAAA------------------------UCGCCUUAGCCCCUUUU-CG---CGAGCGCGAACAAUUGUA-AUUGUAUUUGUAGAGCAGA-AAGGGGUUAA .((((.....------------------------..))))((((((((((.-.(---(....(((((((((....-)))))..))))...))..)-))))))))). ( -24.70) >DroSim_CAF1 41927 100 - 1 UAGGUAAAAACGGUCAGUGAAAUUCGAAAUUUACCCGCCUUAACCCCUUUU-CG---CGAGCGCGAACAAUUGUA-AUUGUAUUUGUAGAGCAGA-AAGGGGUUAA ...........(((..(((((........)))))..))).((((((((((.-.(---(....(((((((((....-)))))..))))...))..)-))))))))). ( -28.10) >DroAna_CAF1 16077 83 - 1 UAGGUACGAAUGGCC-------------------CAGGCCUAACCCCUUUUGUG---CCCACGCUAACAAUUGCAAAUUGUAUUUGUAGAGCGAA-AAGGGGUUAA ...........(((.-------------------...)))(((((((((((((.---...))(((.((((.(((.....))).))))..))).))-))))))))). ( -28.30) >DroPer_CAF1 58260 84 - 1 UAGGUAA---CGG----------------UUCACUCAGCUUAACCCCUUUC-UGGACCGGGCG-GAACAAUUGCAAAUUGUAUUUGUAGAGCGAA-AAGGGGUUAA .......---...----------------...........((((((((((.-((..((....)-).((((.(((.....))).))))....)).)-))))))))). ( -20.80) >consensus UAGGUAAAAACGG________________UU_ACCCGCCUUAACCCCUUUU_CG___CGAGCGCGAACAAUUGCA_AUUGUAUUUGUAGAGCAAA_AAGGGGUUAA ........................................(((((((((.................(((((.....))))).((((.....)))).))))))))). (-15.11 = -14.50 + -0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:16 2006