
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,463,123 – 20,463,232 |
| Length | 109 |
| Max. P | 0.966275 |

| Location | 20,463,123 – 20,463,232 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -13.72 |
| Energy contribution | -15.15 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20463123 109 + 22224390 UAUGACACUGAUCCUAUGUGAAGAUGAACG-----CUUUCACGCACGUCCUAUUUGCGCAUACAAAUUUGCGGACUUAGUCGUGCGUAUAAAGCACGAACG--UAUCUAAAAAUAA ((((.(((.........))).........(-----((((.(((((((..(((..(.((((........)))).)..))).)))))))..))))).....))--))........... ( -27.70) >DroSec_CAF1 36487 109 + 1 UAUCACACUGAUCCUGUGUGAAGAUGAACG-----CUUUCACGCACGUCCUAUUUGCGCAUACAAAUUUGCGAGCUUAGUCGUGCGUAUAAAGCAGGAACG--UAUCUAAAAAUAA ..((((((.......))))))(((((..((-----((((.(((((((..(((....((((........))))....))).)))))))..))))).).....--)))))........ ( -33.40) >DroSim_CAF1 41587 109 + 1 UAUCACACUGAUCCUGUGUGAAGAUGAACG-----CUUUCACGCACGUCCUAUUUGCGCAUACAAAUUUGCGAGCUUGGUCGUGCGUAUAAAGCACGAACG--UAUCUAAAAAUAA ..((((((.......))))))(((((...(-----((((.(((((((.((......((((........)))).....)).)))))))..))))).......--)))))........ ( -35.10) >DroYak_CAF1 35319 95 + 1 UAUGACACUGAUCCUGUGUGAGUAUGCACAUCCUUUUUGCGCAUACAAAUUAUUAGUGCGCAUAAAU--------------GUAU-------CUAAGAGUGUUUAUCUAAAAAUAA ...((((((.....(((((......)))))...(((.(((((((...........))))))).))).--------------....-------.....))))))............. ( -19.30) >consensus UAUCACACUGAUCCUGUGUGAAGAUGAACG_____CUUUCACGCACGUCCUAUUUGCGCAUACAAAUUUGCGAGCUUAGUCGUGCGUAUAAAGCACGAACG__UAUCUAAAAAUAA ..((((((.......))))))(((((((.........)))(((((((.........((((........))))........))))))).................))))........ (-13.72 = -15.15 + 1.44)

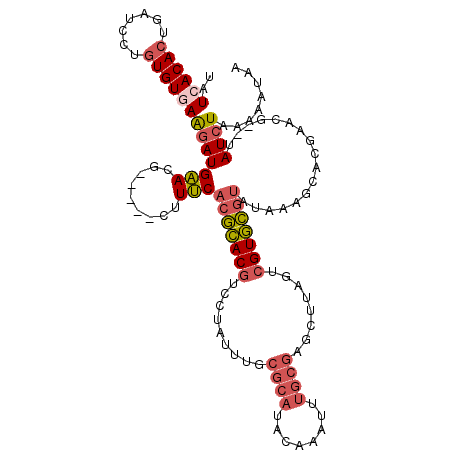
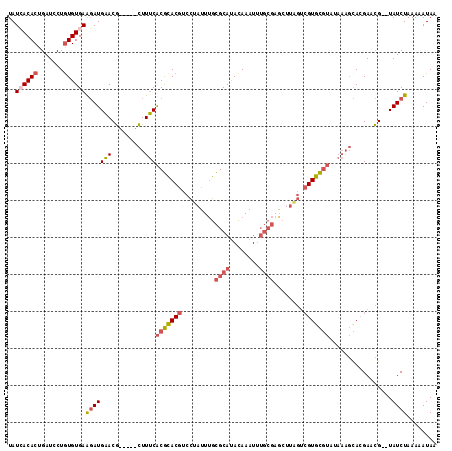
| Location | 20,463,123 – 20,463,232 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -13.50 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20463123 109 - 22224390 UUAUUUUUAGAUA--CGUUCGUGCUUUAUACGCACGACUAAGUCCGCAAAUUUGUAUGCGCAAAUAGGACGUGCGUGAAAG-----CGUUCAUCUUCACAUAGGAUCAGUGUCAUA .........((((--(......(((((.((((((((.(((..(.((((........)))).)..)))..))))))))))))-----)....(((((.....)))))..)))))... ( -31.30) >DroSec_CAF1 36487 109 - 1 UUAUUUUUAGAUA--CGUUCCUGCUUUAUACGCACGACUAAGCUCGCAAAUUUGUAUGCGCAAAUAGGACGUGCGUGAAAG-----CGUUCAUCUUCACACAGGAUCAGUGUGAUA .............--.......(((((.((((((((.(((....((((........))))....)))..))))))))))))-----)........((((((.......)))))).. ( -31.10) >DroSim_CAF1 41587 109 - 1 UUAUUUUUAGAUA--CGUUCGUGCUUUAUACGCACGACCAAGCUCGCAAAUUUGUAUGCGCAAAUAGGACGUGCGUGAAAG-----CGUUCAUCUUCACACAGGAUCAGUGUGAUA ........(((((--(....))(((((.((((((((.((.....((((........))))......)).))))))))))))-----)....))))((((((.......)))))).. ( -31.40) >DroYak_CAF1 35319 95 - 1 UUAUUUUUAGAUAAACACUCUUAG-------AUAC--------------AUUUAUGCGCACUAAUAAUUUGUAUGCGCAAAAAGGAUGUGCAUACUCACACAGGAUCAGUGUCAUA .(((((..(((.......))).))-------))).--------------...((((.(((((........(((((((((.......)))))))))((......))..))))))))) ( -17.70) >consensus UUAUUUUUAGAUA__CGUUCGUGCUUUAUACGCACGACUAAGCUCGCAAAUUUGUAUGCGCAAAUAGGACGUGCGUGAAAG_____CGUUCAUCUUCACACAGGAUCAGUGUCAUA ..................((((((.......)))))).................(((((((.........)))))))..................((((((.......)))))).. (-13.50 = -14.00 + 0.50)

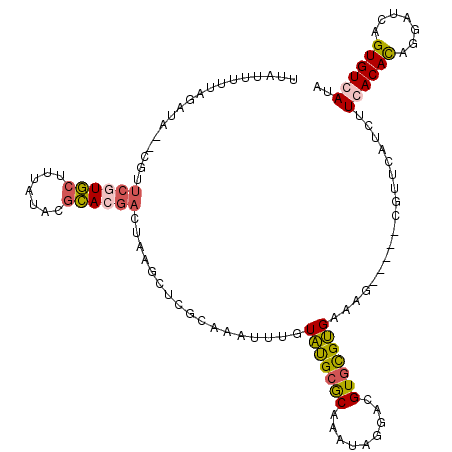
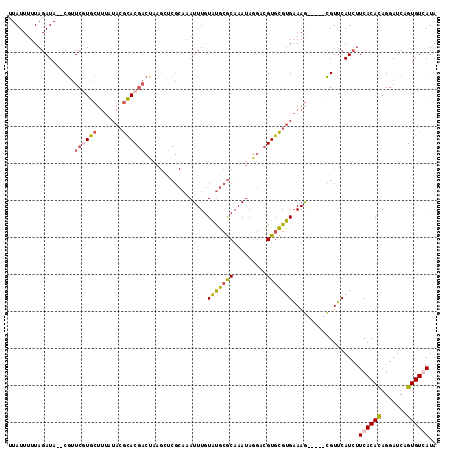
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:14 2006