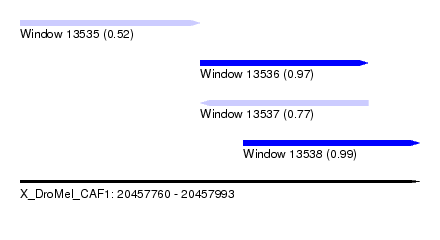
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,457,760 – 20,457,993 |
| Length | 233 |
| Max. P | 0.992406 |

| Location | 20,457,760 – 20,457,865 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.20 |
| Mean single sequence MFE | -35.91 |
| Consensus MFE | -23.42 |
| Energy contribution | -27.04 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20457760 105 + 22224390 AUUGCGUCAAAUUGGCCACCAAAAAUCAAUUGUGGAGAGUUGAAAUCCAUUUCAGGUUAUCCGCGAAUUUCCCUCGCUUGCCGGUGAAAAUCGAGGG-------------CGUAAAAA .(((((((.....(((.............(((((((((.((((((....)))))).)).))))))).............)))(((....)))...))-------------)))))... ( -27.27) >DroSec_CAF1 31534 118 + 1 GUUGCGCCAAAUUGGCCACCGAAAAUCAAUCGUGGAGAGGUGAAAUCCAUUUCAGGUUAUCCGCGAAUUUCCAUCACUUGGCGGUGAAAGUCGAGGGCGGGAAAGGGCGGCGUAAAAA .(((((((......(((............(((((((((((((.....))))))......))))))).(((((....((((((.......))))))....))))).))))))))))... ( -39.50) >DroSim_CAF1 36392 118 + 1 GUUGCGCCAAAUUGCCCACCGAAAAUCAAUUGUGGAGAGGUGAAAUCCAUUUCAGGUUAUCCGCGAAUUUCCAUCGCUUGCCGGGGAAAAUCGAGGGCGGGAAAGGGCGGCGUAAAAA .(((((((.....((((............(((((((((((((.....))))))......))))))).(((((..(((((..(((......))).)))))))))))))))))))))... ( -39.90) >DroEre_CAF1 29075 117 + 1 AACGCGCCAAAAUUGCCACCGA-AAUCAAUUGUGGAGAAGAGAAAUCCAUUUCAGGUUAUCCGUGAAUUUCCAUCGCUUGCCGGUGAAAAUCGAAGGCGGGAAAAGGCGGCGUAAAAA .(((((((........(((.((-((((....(((((.........)))))....)))).)).)))..(((((..(((((..((((....)))).)))))))))).))).))))..... ( -36.30) >DroYak_CAF1 29854 118 + 1 AUUGCGCCAAAUUUGCCACCGAAAAUCAAUUGUGGAGGAGAGAAAUCCAUUUCAGGUUAUCCGCGAAUUUCCAUCGCUUGCCGGUGAAAAUCGAGGGUGGGAAAAGGCGGCGUAAAAA .(((((((...(((.(((((...........(((..(((((((((....)))).((....)).....)))))..)))....((((....))))..))))).)))....)))))))... ( -36.60) >consensus AUUGCGCCAAAUUGGCCACCGAAAAUCAAUUGUGGAGAGGUGAAAUCCAUUUCAGGUUAUCCGCGAAUUUCCAUCGCUUGCCGGUGAAAAUCGAGGGCGGGAAAAGGCGGCGUAAAAA .(((((((.....................(((((((....(((((....))))).....))))))).(((((..(((((..((((....)))).))))))))))....)))))))... (-23.42 = -27.04 + 3.62)



| Location | 20,457,865 – 20,457,963 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 96.54 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -22.00 |
| Energy contribution | -24.56 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.965012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20457865 98 + 22224390 UAAAUACGAAGGGGGAA-AACAUGGGCCUUCGUUGAGAAAUUGCCAGAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGUAGCUGCCUGC .....(((((((.....-........)))))))...........(((.((.....))......((((((........(((......)))))))))))). ( -18.92) >DroSec_CAF1 31652 98 + 1 UAAAUACGAAGGGGGAA-AACAUGGGCCUUCGGUGAGAAAUUGCCGAAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGUAGCUGCCUGC ..........(.(((..-....(((((((((((..(....)..)))))))....))))....(((.(((((.............))))).))).))).) ( -28.12) >DroSim_CAF1 36510 98 + 1 UAAAUACAAAGGGGGAA-AACAUGGGCCUUCGGUGAGAAAUUGCCGAAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGUAGCUGCCUGC ..........(.(((..-....(((((((((((..(....)..)))))))....))))....(((.(((((.............))))).))).))).) ( -28.12) >DroEre_CAF1 29192 98 + 1 UAAAUACAAAGGGGGAA-AACAUGGGCCUUCGGUGAGAAAUUGCCGAAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGAAGCUGCCUGC ........(((..(((.-........(((((((..(....)..)))))))....))).)))..((((((((................)))))))).... ( -29.81) >DroYak_CAF1 29972 99 + 1 UAAAUACAAAGGGGGAAAAACGUGGGCCUUCGGUGAGAAAUUGCCGGAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGAAGCUGCCUGC ..........(.((.......((((((((((((..(....)..)))))))....)))))....((((((((................)))))))))).) ( -32.19) >consensus UAAAUACAAAGGGGGAA_AACAUGGGCCUUCGGUGAGAAAUUGCCGAAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGUAGCUGCCUGC ..........(.(((.......((((((((((((((....))))))))))....))))....(((.(((((.............))))).))).))).) (-22.00 = -24.56 + 2.56)

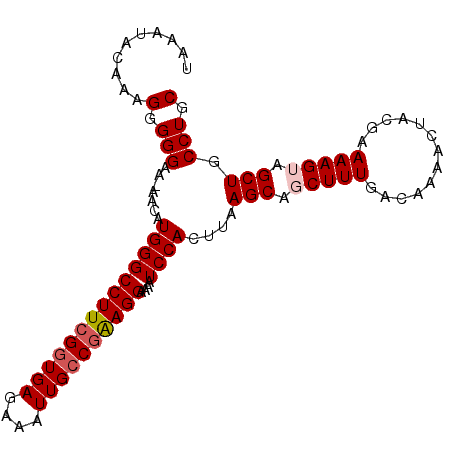
| Location | 20,457,865 – 20,457,963 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 96.54 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -19.96 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20457865 98 - 22224390 GCAGGCAGCUACUUUUCGUAGUUUUGUCAAAGCUGCUUAAGUGGAUUUUCCUCUGGCAAUUUCUCAACGAAGGCCCAUGUU-UUCCCCCUUCGUAUUUA (((((((((((((......)))........)))))))).((.((.....)).)).)).........(((((((........-.....)))))))..... ( -22.92) >DroSec_CAF1 31652 98 - 1 GCAGGCAGCUACUUUUCGUAGUUUUGUCAAAGCUGCUUAAGUGGAUUUUCCUUCGGCAAUUUCUCACCGAAGGCCCAUGUU-UUCCCCCUUCGUAUUUA ...(((..((((((...(((((((.....)))))))..))))))......((((((..........)))))))))......-................. ( -23.70) >DroSim_CAF1 36510 98 - 1 GCAGGCAGCUACUUUUCGUAGUUUUGUCAAAGCUGCUUAAGUGGAUUUUCCUUCGGCAAUUUCUCACCGAAGGCCCAUGUU-UUCCCCCUUUGUAUUUA ...(((..((((((...(((((((.....)))))))..))))))......((((((..........)))))))))......-................. ( -23.70) >DroEre_CAF1 29192 98 - 1 GCAGGCAGCUUCUUUUCGUAGUUUUGUCAAAGCUGCUUAAGUGGAUUUUCCUUCGGCAAUUUCUCACCGAAGGCCCAUGUU-UUCCCCCUUUGUAUUUA ..(((((((((.....((......))...)))))))))..((((.....(((((((..........))))))).))))...-................. ( -23.30) >DroYak_CAF1 29972 99 - 1 GCAGGCAGCUUCUUUUCGUAGUUUUGUCAAAGCUGCUUAAGUGGAUUUUCCUCCGGCAAUUUCUCACCGAAGGCCCACGUUUUUCCCCCUUUGUAUUUA ..(((((((((.....((......))...)))))))))..((((.....(((.(((..........))).))).))))..................... ( -21.10) >consensus GCAGGCAGCUACUUUUCGUAGUUUUGUCAAAGCUGCUUAAGUGGAUUUUCCUUCGGCAAUUUCUCACCGAAGGCCCAUGUU_UUCCCCCUUUGUAUUUA ..((((((((......((......))....))))))))..((((.....(((((((..........))))))).))))..................... (-19.96 = -20.24 + 0.28)


| Location | 20,457,890 – 20,457,993 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.09 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -23.35 |
| Energy contribution | -25.51 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20457890 103 + 22224390 CCUUCGUUGAGAAAUUGCCAGAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGUAGCUGCCUGCCGGGAAAUCAACUCCGGAGAUACAGAUACA---------- .(((((((((....(((.(((.((.....))......((((((........(((......)))))))))))).)))....)))))...))))...........---------- ( -21.20) >DroSec_CAF1 31677 103 + 1 CCUUCGGUGAGAAAUUGCCGAAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGUAGCUGCCUGCCGGGAAAUCAACUUCGAAGAUACAGAUACA---------- (((((((..(....)..)))))))....(((......((((((........(((......)))))))))......))).........................---------- ( -28.40) >DroSim_CAF1 36535 103 + 1 CCUUCGGUGAGAAAUUGCCGAAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGUAGCUGCCUGCCGGGAAAUCAACUUCGAAGAUACAGAUACA---------- (((((((..(....)..)))))))....(((......((((((........(((......)))))))))......))).........................---------- ( -28.40) >DroEre_CAF1 29217 113 + 1 CCUUCGGUGAGAAAUUGCCGAAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGAAGCUGCCUGCCGGGAAAUCAACUCCGUAGAUACAGAUACAGAUACAGAUA (((((((..(....)..)))))))...(((.......((((((((................))))))))((((.(((.......))).)))).....)))............. ( -32.79) >DroYak_CAF1 29998 113 + 1 CCUUCGGUGAGAAAUUGCCGGAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGAAGCUGCCUGCCGGGAAAUCAACGCCGGAGAUACAGAUACAGAUACAGAUA (((((((..(....)..)))))))...(((.......((((((((................))))))))((.((((..........)))))).....)))............. ( -32.89) >consensus CCUUCGGUGAGAAAUUGCCGAAGGAAAAUCCACUUAAGCAGCUUUGACAAAACUACGAAAAGUAGCUGCCUGCCGGGAAAUCAACUCCGAAGAUACAGAUACA__________ ((((((((((....))))))))))....(((......((((((....................))))))......)))................................... (-23.35 = -25.51 + 2.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:10 2006