| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,306,134 – 2,306,241 |
| Length | 107 |
| Max. P | 0.561793 |

| Location | 2,306,134 – 2,306,241 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -20.56 |
| Energy contribution | -20.42 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
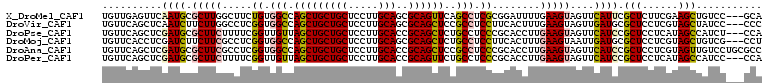
>X_DroMel_CAF1 2306134 107 - 22224390 UGUUGAGUUCAAUGCGCUUGGCUUCUGUGGCCAGCUGCUGCUCCUUGCAGCGCAGUUCAGCCUCGCGGAUUUUGAAGUAGUUCAUUCGCUCUUCGAAGCUGUCC---GCA .((((((.....((((((.((((.....))))))).(((((.....)))))))).))))))...(((((((((((((.(((......))))))))))...))))---)). ( -43.60) >DroVir_CAF1 19327 107 - 1 UGUUCAGCUCAAUCUUCUUGGCCUCGGUGGCCAGCUGCUGCUCCUUGCAGCGCAGCUCCGCCUCCUUCACUUUGAAGUAGUUGAUGCGCUCCUCGUAGCUAUCC---CCC .((((((((....((((..((....((.(((.((((((.(((......)))))))))..))).))....))..)))).)))))).))(((......))).....---... ( -32.80) >DroPse_CAF1 6918 107 - 1 UGUUCAGCUCGAUGCGCUUCUUUUCGGUUGUUAGCUGCUGCUCCUUGCACCGCAGCUCUGCCUCCCGCACCUUGAAGUAGUUCAUCCGCUCCUCAUAGCCAUCU---CCA ((...(((..((((.(((.((((..(((....(((((((((.....)))..))))))..((.....)))))..)))).))).)))).)))...)).........---... ( -27.60) >DroMoj_CAF1 9181 107 - 1 UGUUCACCUCGAUCUUCUUCGCCUCGGUGGCCAGCUGCUGCUCCUUGCAGCGCAGCUCUGCCUCCUUCACUUUGAAGUAAUUGAUGCGCUCCUCGUAGCUGUCG---CCU .........(((......)))....((((((.(((((((((.....)))(((((..((......((((.....)))).....))))))).....))))))))))---)). ( -32.80) >DroAna_CAF1 5416 110 - 1 UGUUCAGCUCGAUGCGCUUCGCCUCGGUGGCCAGCUGCUGCUCCUUGCACCGCAGCUCCGCCUCCCGCACCUUGAAGUAGUUCAUCCGCUCCUCGUAGUUGUCCUGCGCC .(..((((((((.(.((...((...(((((..(((((((((.....)))..)))))))))))....))....((((....))))...)).).))).)))))..)...... ( -33.70) >DroPer_CAF1 9491 107 - 1 UGUUCAGCUCGAUGCGCUUCUUUUCGGUUGUUAGCUGCUGCUCCUUGCACCGCAGUUCUGCCUCCCGCACCUUGAAGUAGUUCAUCCGCUCCUCAUAGCCAUCC---CCA ((...(((..((((.(((.((((..(((....(((((((((.....)))..))))))..((.....)))))..)))).))).)))).)))...)).........---... ( -25.20) >consensus UGUUCAGCUCGAUGCGCUUCGCCUCGGUGGCCAGCUGCUGCUCCUUGCACCGCAGCUCUGCCUCCCGCACCUUGAAGUAGUUCAUCCGCUCCUCGUAGCUAUCC___CCA ..........((((.(((((.....((.(((.(((((((((.....)))..))))))..))).))........)))))....)))).(((......)))........... (-20.56 = -20.42 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:41 2006