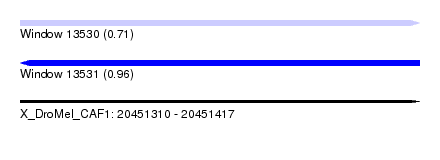
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,451,310 – 20,451,417 |
| Length | 107 |
| Max. P | 0.959302 |

| Location | 20,451,310 – 20,451,417 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.55 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -14.87 |
| Energy contribution | -16.32 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20451310 107 + 22224390 AAAAA----U----CGUGGAGCUGCGGCAUCUUCUGCUUUUGACACAAGUUAAUGAGUUUCGCGCUUUUGCGCCUGCCAAAUGCCACCAGUGGGUGUACUUAAAACUCCAACCAA .....----.----..(((((....((((......(((((((((....))))).))))...((((....)))).))))......((((....)))).........)))))..... ( -27.90) >DroVir_CAF1 41487 95 + 1 ---------UUUCUUGGGGAGC----------UCUGCGCCUGACACAAGUUAAUGAGUUUCGCGCUUUUGCGCCAGCCAAAUGCCACCAUCGGGUGCACUUAAUU-CCCAACAAC ---------....(((((((..----------..(((((((((.....((...((.(((..((((....)))).)))))...)).....))))))))).....))-))))).... ( -32.10) >DroPse_CAF1 43290 103 + 1 -ACAGCCCCUG---GGCUGUGCUGUGGCAUCUUCUGCUCUUGACACAAGUUAAUGAGUUUCGCGCUUUCGCGCCUGCCAAAUGCCACCAUCGGGUGUACUUAACGCU-------- -((((((....---))))))((..(((((......(((((((((....))))).))))...((((....)))).)))))...))((((....))))...........-------- ( -36.90) >DroYak_CAF1 22900 107 + 1 -AAAA----UU---GCUGGAGCUGCGGCCUCUUCUGCUUUUGACACAAGUUAAUGAGUUUCGCGCUUUUGCGCCUGCCAAAUGCCACCAGCGGGUGUACUUAAAACUCCAACCAC -....----.(---(((((.((...(((.......(((((((((....))))).))))...((((....))))..)))....))..))))))((.((.......)).))...... ( -27.60) >DroMoj_CAF1 55296 100 + 1 GCCCA----UUUCUUGGGGAGC----------UCUGCGCCUGACACAAGUUAAUGAGUUUCGCGCUUUCGCGCCAGCCAAAUGCCACCAUCGGGUGCACUUAAUU-UCCAACAAC .....----....((((..(..----------..(((((((((.....((...((.(((..((((....)))).)))))...)).....))))))))).....).-.)))).... ( -28.90) >DroPer_CAF1 43757 103 + 1 -ACAGCCCGUG---GGCUGUGCUGUGGCAUCUUCUGCUCUUGACACAAGUUAAUGAGUUUCGCGCUUUUGCGCUUGCCAAAUGCCACCAUCGGGUGUACUUAACGCU-------- -((((((....---))))))((..(((((......(((((((((....))))).))))...((((....)))).)))))...))((((....))))...........-------- ( -38.00) >consensus _ACAA____UU___GGCGGAGCUG_GGCAUCUUCUGCUCUUGACACAAGUUAAUGAGUUUCGCGCUUUUGCGCCUGCCAAAUGCCACCAUCGGGUGUACUUAAAACUCCAAC_AC ....................((...(((.......(((((((((....))))).))))...((((....))))..)))....))((((....))))................... (-14.87 = -16.32 + 1.44)

| Location | 20,451,310 – 20,451,417 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.55 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -21.35 |
| Energy contribution | -22.38 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20451310 107 - 22224390 UUGGUUGGAGUUUUAAGUACACCCACUGGUGGCAUUUGGCAGGCGCAAAAGCGCGAAACUCAUUAACUUGUGUCAAAAGCAGAAGAUGCCGCAGCUCCACG----A----UUUUU .....(((((((...(((......))).(((((((((.((..((((....))))((.((..........)).))....))...))))))))))))))))..----.----..... ( -32.30) >DroVir_CAF1 41487 95 - 1 GUUGUUGGG-AAUUAAGUGCACCCGAUGGUGGCAUUUGGCUGGCGCAAAAGCGCGAAACUCAUUAACUUGUGUCAGGCGCAGA----------GCUCCCCAAGAAA--------- .((.(((((-...((((((((((....))).)))))))(((.((((....((((((...........))))))...))))..)----------))..))))).)).--------- ( -32.10) >DroPse_CAF1 43290 103 - 1 --------AGCGUUAAGUACACCCGAUGGUGGCAUUUGGCAGGCGCGAAAGCGCGAAACUCAUUAACUUGUGUCAAGAGCAGAAGAUGCCACAGCACAGCC---CAGGGGCUGU- --------.((((.....))........(((((((((.((..((((....))))............((((...)))).))...))))))))).))((((((---....))))))- ( -41.00) >DroYak_CAF1 22900 107 - 1 GUGGUUGGAGUUUUAAGUACACCCGCUGGUGGCAUUUGGCAGGCGCAAAAGCGCGAAACUCAUUAACUUGUGUCAAAAGCAGAAGAGGCCGCAGCUCCAGC---AA----UUUU- ...(((((((((....((.((((....))))))....(((..((((....))))............(((.(((.....))).)))..)))..)))))))))---..----....- ( -34.50) >DroMoj_CAF1 55296 100 - 1 GUUGUUGGA-AAUUAAGUGCACCCGAUGGUGGCAUUUGGCUGGCGCGAAAGCGCGAAACUCAUUAACUUGUGUCAGGCGCAGA----------GCUCCCCAAGAAA----UGGGC .((.((((.-...((((((((((....))).)))))))(((.((((....)))).............(((((.....))))))----------))...)))).)).----..... ( -30.60) >DroPer_CAF1 43757 103 - 1 --------AGCGUUAAGUACACCCGAUGGUGGCAUUUGGCAAGCGCAAAAGCGCGAAACUCAUUAACUUGUGUCAAGAGCAGAAGAUGCCACAGCACAGCC---CACGGGCUGU- --------.((((.....))........(((((((((.((..((((....))))............((((...)))).))...))))))))).))((((((---....))))))- ( -37.50) >consensus GU_GUUGGAGAAUUAAGUACACCCGAUGGUGGCAUUUGGCAGGCGCAAAAGCGCGAAACUCAUUAACUUGUGUCAAGAGCAGAAGAUGCC_CAGCUCCACC___AA____UUGU_ ......(((((........((((....))))((.((((((..((((....))))(.....)..........)))))).)).............)))))................. (-21.35 = -22.38 + 1.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:04 2006