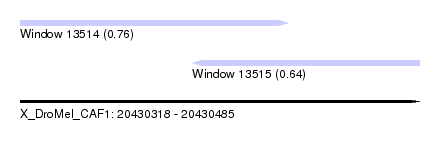
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,430,318 – 20,430,485 |
| Length | 167 |
| Max. P | 0.763545 |

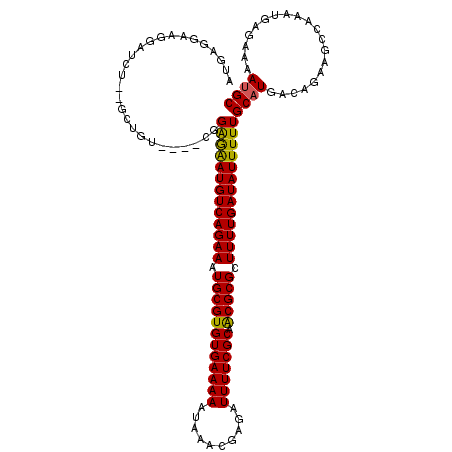
| Location | 20,430,318 – 20,430,430 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.51 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -24.77 |
| Energy contribution | -24.30 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
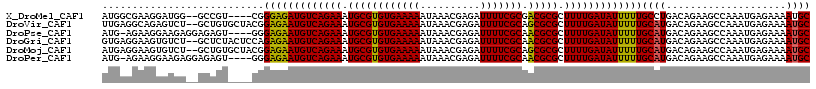
>X_DroMel_CAF1 20430318 112 + 22224390 AUGGCGAAGGAUGG--GCCGU----CGGGAGAUGUCAGAAAUGCGUGUGAAAAAUAAACGAGAUUUUCGCGACGCGCUUUUGAUAUUUUUGCCUGACAGAAGCCAAAUGAGAAAAUGC .((((...((....--.))((----((((.((((((((((.((((((((((((..........))))))).))))).))))))))))....))))))....))))............. ( -39.00) >DroVir_CAF1 18535 116 + 1 UUGAGGCAGAGUCU--GCUGUGCUACGGAGAAUGUCAGAAAUGCGUGUGAAAAAUAAACGAGAUUUUCGCAGCGCGCUUUUGAUAUUUUUGCAUGACAGAAGCCAAAUGAGAAAAUGC ....(((....(((--(..((((....(((((((((((((.((((((((((((..........)))))))).)))).)))))))))))))))))..)))).))).............. ( -38.50) >DroPse_CAF1 18961 113 + 1 AUG-AGAAGGAAGAGGAGAGU----GGGAGAAUGUCAGAAAUGCGUGUGAAAAAUAAACGAGAUUUUCGCAACGCGCUUUUGAUAUUUUUGCAUGACAGAAGCCAAAUGAGAAAAUGC ...-....((.........((----(.(((((((((((((.((((((((((((..........))))))).))))).))))))))))))).)))........)).............. ( -31.33) >DroGri_CAF1 16987 116 + 1 GUGAGGAAGUGUCU--GCUCUACUCCAGAGAAUGUCAGAAAUGCGUGUGAAAAAUAAACGAGAUUUUCGCAACGCGCUUUUGAUAUUUUUGCAUGACAGAAGCCAAAUGAGAAAAUGC ....((...(((((--((.........(((((((((((((.((((((((((((..........))))))).))))).)))))))))))))))).))))....)).............. ( -31.80) >DroMoj_CAF1 27473 116 + 1 AUGAGGAAGUGUCU--GCUGUGCUACGGAGAAUGUCAGAAAUGCGUGUGAAAAAUAAACGAGAUUUUCGCAGCGCGCUUUUGAUAUUUUUGCAUGACAGAAGCCAAAUGAGAAAAUGC ....((.....(((--(..((((....(((((((((((((.((((((((((((..........)))))))).)))).)))))))))))))))))..))))..)).............. ( -35.80) >DroPer_CAF1 19888 113 + 1 AUG-AGAAGGAAGAGGAGAGU----GGGAGAAUGUCAGAAAUGCGUGUGAAAAAUAAACGAGAUUUUCGCAACGCGCUUUUGAUAUUUUUGCAUGACAGAAGCCAAAUGAGAAAAUGC ...-....((.........((----(.(((((((((((((.((((((((((((..........))))))).))))).))))))))))))).)))........)).............. ( -31.33) >consensus AUGAGGAAGGAUCU__GCUGU____CGGAGAAUGUCAGAAAUGCGUGUGAAAAAUAAACGAGAUUUUCGCAACGCGCUUUUGAUAUUUUUGCAUGACAGAAGCCAAAUGAGAAAAUGC ...........................(((((((((((((.((((((((((((..........))))))).))))).)))))))))))))((((....................)))) (-24.77 = -24.30 + -0.47)

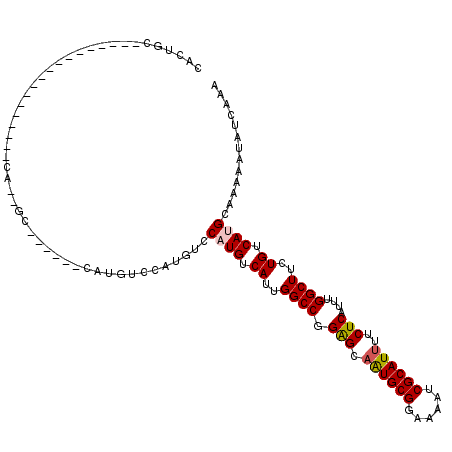
| Location | 20,430,390 – 20,430,485 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -18.91 |
| Energy contribution | -19.05 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20430390 95 - 22224390 CACGUC------------------CACGUC------CAUGUCCAUGUCCAUGUCAUUGGCCGGGGCAAUGCGGAAAAUCGCAUUUUCUCAUUUGGCUUCUGUCAGGCAAAAAUAUCAAA ...(((------------------...(..------(((....)))..).((.((..((((((((.((((((......)))))).))))....))))..)).)))))............ ( -22.40) >DroVir_CAF1 18611 92 - 1 CAUUGC------------------C---GG------CAUAUCCAUGCUCAUGUCAUUGGCCGGAGCUGUGCGGAAAAUCGCAUUUUCUCAUUUGGCUUCUGUCAUGCAAAAAUAUCAAA ..((((------------------.---((------(((....))))).(((.((..((((.(((..(((((......)))))...)))....))))..)).))))))).......... ( -25.30) >DroPse_CAF1 19034 95 - 1 CAUUGC------------------CAUUGC------CACCGCCAUGUCCAUGUCAUUGGCCGGAGCAAUGCGGAAAAUCGCAUUUUCUCAUUUGGCUUCUGUCAUGCAAAAAUAUCAAA ..((((------------------(((.((------....)).)))...(((.((..((((.(((.((((((......))))))..)))....))))..)).))))))).......... ( -23.60) >DroGri_CAF1 17063 92 - 1 CAUUGC------------------C---GC------CAUGUUCAUGCUCAUGUCAUUGGCCGGAGCAAUGCGGAAAAUCGCAUUUUCUCAUUUGGCUUCUGUCAUGCAAAAAUAUCAAA ......------------------.---..------.(((((..(((..(((.((..((((.(((.((((((......))))))..)))....))))..)).))))))..))))).... ( -22.40) >DroEre_CAF1 2558 119 - 1 CACGUCCACGUCCACGUUCACGUCCACAUCCACGUCCAUGUCCAUGGCCAUGUCAUUGGCCGGGGCAAUGCGGAAAAUCGCAUUUUCUCAUUUGGCUUCUGUCAGGCAAAAAUAUCAAA .((((..(((....)))..))))..........(((..(((((.((((((......)))))))))))(((((......))))).........((((....)))))))............ ( -32.00) >DroPer_CAF1 19961 95 - 1 CACUGC------------------CAUUGC------UACCGCCAUGUCCAUGUCAUUGGCCGGAGCAAUGCGGAAAAUCGCAUUUUCUCAUUUGGCUUCUGUCAUGCAAAAAUAUCAAA ..((((------------------.(((((------(.((((((((.......)).))).)))))))))))))......(((..........((((....)))))))............ ( -24.80) >consensus CACUGC__________________CA__GC______CAUGUCCAUGUCCAUGUCAUUGGCCGGAGCAAUGCGGAAAAUCGCAUUUUCUCAUUUGGCUUCUGUCAUGCAAAAAUAUCAAA ................................................((((.((..((((.(((.((((((......))))))..)))....))))..)).))))............. (-18.91 = -19.05 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:50 2006