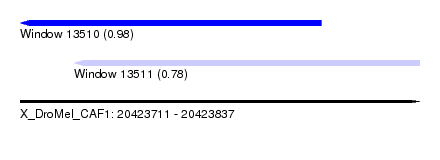
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,423,711 – 20,423,837 |
| Length | 126 |
| Max. P | 0.976833 |

| Location | 20,423,711 – 20,423,806 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -29.37 |
| Energy contribution | -29.60 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20423711 95 - 22224390 GCAACGGCGCAUUACACUGAGGGCAAAGUGGCCAAGUGAGUGAAAACUCAGGUCGAGUGAAAAGCGUUGCUAAAAAGCGAGGCUAAAACCACUCG .....(((.......(((...(((......))).)))((((....))))..)))(((((...(((.(((((....))))).))).....))))). ( -31.50) >DroSec_CAF1 12888 95 - 1 GCAACGGCGCAUUACACUGAGGGCAAAGUGGCCAAGUGAGUGAAAACUCAGGCCGAGUGAAAAGCGUUGCUAAAAAGCGAGGCUAAAACCACUCA .....(((.......(((...(((......))).)))((((....))))..)))(((((...(((.(((((....))))).))).....))))). ( -34.20) >DroYak_CAF1 9171 95 - 1 GCAACGGCGCAUUACACUGAGGCCAAAGUGGCCAAGUGAGUGAAAACUCGGGCCGAGUGAAAAGCGUGACUGUAAAGCGAAGCUAAAAGCACUCA .....(((.(.....(((..((((.....)))).)))((((....))))).)))(((((...(((....((....))....))).....))))). ( -32.80) >consensus GCAACGGCGCAUUACACUGAGGGCAAAGUGGCCAAGUGAGUGAAAACUCAGGCCGAGUGAAAAGCGUUGCUAAAAAGCGAGGCUAAAACCACUCA .....(((......((((........))))......(((((....))))).)))(((((...(((.(((((....))))).))).....))))). (-29.37 = -29.60 + 0.23)


| Location | 20,423,728 – 20,423,837 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -34.39 |
| Consensus MFE | -23.68 |
| Energy contribution | -24.16 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20423728 109 - 22224390 ---------UGCGAGUCCUUAAUUUGCUAAUUGAGAGCCUGCAACGGCGCAUUACACUGAGGGCAAAGUGGCC-AAGUGAGUGAAAACUCAG-GUCGAGUGAAAAGCGUUGCUAAAAAGC ---------((((.(..(((((((....)))))))..).)))).((((((.((.((((((.(((......)))-...(((((....))))).-.)).)))).)).))))))......... ( -34.40) >DroSec_CAF1 12905 109 - 1 ---------UGCGAGUCCUUAAUUUGCUAAUUGAGAGCCUGCAACGGCGCAUUACACUGAGGGCAAAGUGGCC-AAGUGAGUGAAAACUCAG-GCCGAGUGAAAAGCGUUGCUAAAAAGC ---------((((.(..(((((((....)))))))..).)))).((((((.(((((((........)))((((-...(((((....))))))-)))..))))...))))))......... ( -34.80) >DroAna_CAF1 16184 103 - 1 ---------UG-GAGUGCUUAAUUUGCUAAUUGAGAGCCUGCAACGGCGCAUAACACUAAAGACCAAGUGCCAGCAGCGAGUGAGAACUCAAAAGCGAGUGGA-------GCUCAAAAUG ---------..-..(.((((.(((((((........((.(((...(((((.................))))).)))))((((....))))...))))))).))-------)).)...... ( -29.33) >DroEre_CAF1 12250 118 - 1 GAGUGCGAGUGCGAGUCCUUAAUUUGCUAAUUGAGAGCCUGCAACGGCGCAUUACACUGAGGGCAAAGUGGGC-AAGUGAGUGGAAACUCAG-GCCGAGCGAAAAGCGUGACUAAAAAGC .((((((..((((.(..(((((((....)))))))..).))))....)))))).((((........))))(((-...(((((....))))).-)))..((.....))............. ( -37.40) >DroYak_CAF1 9188 112 - 1 ------GAGUGCGAGUCCUUAAUUUGCUAAUUGAGAGCCUGCAACGGCGCAUUACACUGAGGCCAAAGUGGCC-AAGUGAGUGAAAACUCGG-GCCGAGUGAAAAGCGUGACUGUAAAGC ------...((((.(..(((((((....)))))))..).))))(((((((....((((..((((.....))))-.)))).((....(((((.-..))))).....))))).))))..... ( -36.00) >consensus _________UGCGAGUCCUUAAUUUGCUAAUUGAGAGCCUGCAACGGCGCAUUACACUGAGGGCAAAGUGGCC_AAGUGAGUGAAAACUCAG_GCCGAGUGAAAAGCGUGGCUAAAAAGC .........((((.(..(((((((....)))))))..).)))).((((......((((........))))........((((....))))...))))....................... (-23.68 = -24.16 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:47 2006