
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,422,368 – 20,422,473 |
| Length | 105 |
| Max. P | 0.734753 |

| Location | 20,422,368 – 20,422,473 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -22.95 |
| Energy contribution | -22.95 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
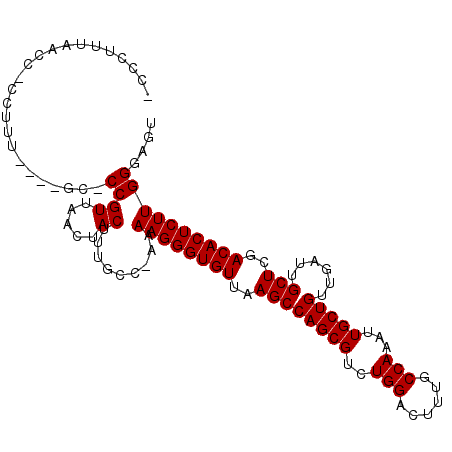
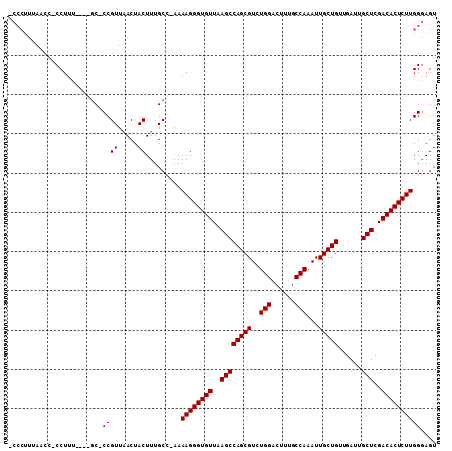
>X_DroMel_CAF1 20422368 105 + 22224390 -CCCUUUAACC-CCUUUUUGCCC-CCGUUAACUACUUUGCA-AAAAGGGUGUUAAGCCAGCGUCUGGACUUUGCCAAAUUGCUGUUGAUUGCUCGACACUCUUGGGAGU -(((.......-(((((((((..-..((.....))...)))-))))))((((..((((((((..(((......)))...)))))......)))..))))....)))... ( -31.70) >DroSec_CAF1 11554 101 + 1 -CCCUUUAACC-CCUUU----GC-CCGUUAACUACUUUGCC-AAAAGGGUGUUAAGCCAGCGUCUGGACUUUGCCAAAUUGCUGUUGAUUGCUCGACACUCUUGGGAGU -(((.(((((.-.....----..-..)))))..........-..((((((((..((((((((..(((......)))...)))))......)))..)))))))))))... ( -26.70) >DroEre_CAF1 10938 104 + 1 ACCCUUUAACUCCCUUC----GC-CCGUUAACUACUUUGCCAAAAAGGGUGUUAAGCCAGCGUCUGGACUUUGCCAAAUUGCUGUUGAUUGCUCGACACUCUUGGGAGU ........((((((...----((-..((.....))...))....((((((((..((((((((..(((......)))...)))))......)))..)))))))))))))) ( -31.80) >DroYak_CAF1 7828 93 + 1 -----------CCCUUG----GCCCCGUUGACUACUUUGCG-AAAAGGGUGUUAAGCCAGCGUCUGGACUUUGCCAAAUUGCUGUUGAUUGCUCGACACUCUUGGCAGU -----------......----(((.(((..........)))-..((((((((..((((((((..(((......)))...)))))......)))..)))))))))))... ( -27.70) >consensus _CCCUUUAACC_CCUUU____GC_CCGUUAACUACUUUGCC_AAAAGGGUGUUAAGCCAGCGUCUGGACUUUGCCAAAUUGCUGUUGAUUGCUCGACACUCUUGGGAGU ........................((((.....)).........((((((((..((((((((..(((......)))...)))))......)))..)))))))))).... (-22.95 = -22.95 + -0.00)

| Location | 20,422,368 – 20,422,473 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -20.35 |
| Energy contribution | -21.48 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
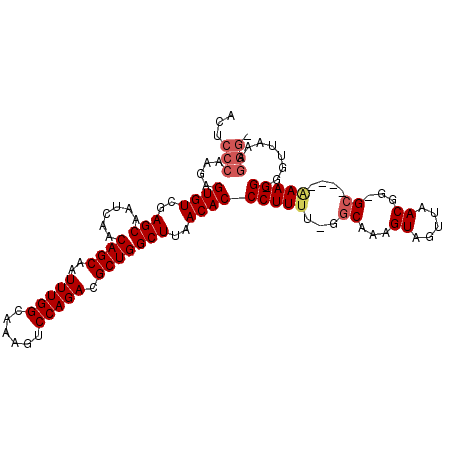
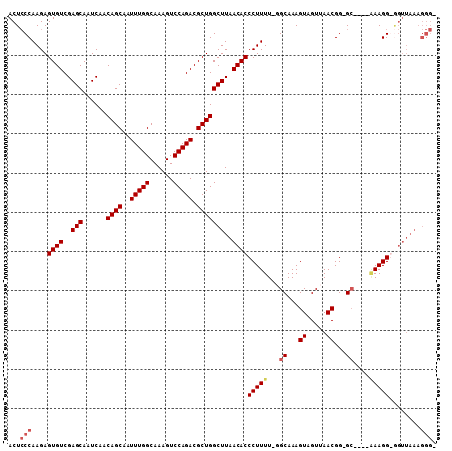
>X_DroMel_CAF1 20422368 105 - 22224390 ACUCCCAAGAGUGUCGAGCAAUCAACAGCAAUUUGGCAAAGUCCAGACGCUGGCUUAACACCCUUUU-UGCAAAGUAGUUAACGG-GGGCAAAAAGG-GGUUAAAGGG- ...(((...((((((((....))....((......))........))))))...(((((.(((((((-(((...((.....))..-..)))))))))-)))))).)))- ( -34.80) >DroSec_CAF1 11554 101 - 1 ACUCCCAAGAGUGUCGAGCAAUCAACAGCAAUUUGGCAAAGUCCAGACGCUGGCUUAACACCCUUUU-GGCAAAGUAGUUAACGG-GC----AAAGG-GGUUAAAGGG- ...(((..((((...((....))..((((..(((((......))))).))))))))...((((((((-(.(...((.....)).)-.)----)))))-)))....)))- ( -30.50) >DroEre_CAF1 10938 104 - 1 ACUCCCAAGAGUGUCGAGCAAUCAACAGCAAUUUGGCAAAGUCCAGACGCUGGCUUAACACCCUUUUUGGCAAAGUAGUUAACGG-GC----GAAGGGAGUUAAAGGGU (((((((((.((((..(((......((((..(((((......))))).)))))))..)))).))).((.((...((.....))..-))----.))))))))........ ( -30.60) >DroYak_CAF1 7828 93 - 1 ACUGCCAAGAGUGUCGAGCAAUCAACAGCAAUUUGGCAAAGUCCAGACGCUGGCUUAACACCCUUUU-CGCAAAGUAGUCAACGGGGC----CAAGGG----------- ..(((((((..(((.((....)).)))....)))))))(((.((((...)))))))....(((((..-.((...((.....))...))----.)))))----------- ( -23.30) >consensus ACUCCCAAGAGUGUCGAGCAAUCAACAGCAAUUUGGCAAAGUCCAGACGCUGGCUUAACACCCUUUU_GGCAAAGUAGUUAACGG_GC____AAAGG_GGUUAAAGGG_ ...(((....((((..(((......((((..(((((......))))).)))))))..))))(((((...((...((.....))...))....)))))........))). (-20.35 = -21.48 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:44 2006