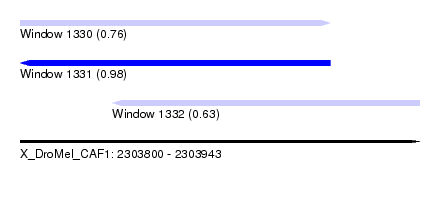
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,303,800 – 2,303,943 |
| Length | 143 |
| Max. P | 0.983906 |

| Location | 2,303,800 – 2,303,911 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -19.98 |
| Energy contribution | -19.14 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
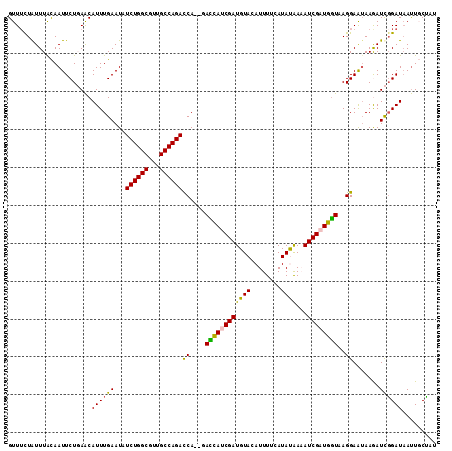
>X_DroMel_CAF1 2303800 111 + 22224390 GUUAUUAUUUACAAUUCUGAACAUUUGAAUAUCUGGCGUUGCCAGACCA--GAUCAUCGAUGUACAUUUUCAUAUAAAAUCGAUGAUAAGGAAUAAGAUCGGAUAAUUGCUAU ...........(((((((((.(.........((((((...))))))((.--.(((((((((.................)))))))))..)).....).)))))..)))).... ( -26.83) >DroSec_CAF1 1787 110 + 1 GUUUCUGUUUACAAUUCUGAACAUUUGAAUAUCUGGCGUUGCCAGACCA--GACCACCGAUGUACAUUUUCAUAUAA-AUCGAUGGUAAGGAAUAAGAUCGGAUAAUUGCUAU ...........(((((((((.(.........((((((...))))))((.--.((((.((((................-)))).))))..)).....).)))))..)))).... ( -24.29) >DroSim_CAF1 2590 110 + 1 GUUUCUGUUUACAAUUCUGAACAUUUGAAUAUCUGGCGUUGCCAGACCA--GACCACCGAUGUAGAUUUUCAUAUAA-AUCGAUGGUAAGGAAUAAGAUCGGAUAAUUGCUAU ...........(((((((((.(.........((((((...))))))((.--.((((.(((((((........)))..-)))).))))..)).....).)))))..)))).... ( -24.30) >DroEre_CAF1 2932 113 + 1 GUUUCUAUUUACAAUUCUUAACAUUUGAAUCUCUGGCGUUGCCAGAUCAACUACCAUCGAUGUACAUUUUUAUACGAAAUCGAUGGUAAGGAAAAAAAUCGGAUAAUUUCCUU .(((((..................((((...((((((...)))))))))).((((((((((.................)))))))))).)))))......(((.....))).. ( -27.43) >DroYak_CAF1 2638 113 + 1 GUUUCUAUUUGUAAUUCUUAGCAUUUGAAUCUCUGGCGUUGCCAGAUCAACGAUUAUCGAUGUACAUUUUAAUACGAAAUCGAUGGUAAGGAGUAAAAUUUGAUCACUUCAUU ...((.((((....((((((.(((((((...((((((...))))))))))(((((.(((.(((........)))))))))))))).))))))...))))..)).......... ( -23.40) >consensus GUUUCUAUUUACAAUUCUGAACAUUUGAAUAUCUGGCGUUGCCAGACCA__GACCAUCGAUGUACAUUUUCAUAUAAAAUCGAUGGUAAGGAAUAAGAUCGGAUAAUUGCUAU ......................((((((...((((((...))))))((....((((((((((((........))))...))))))))..)).......))))))......... (-19.98 = -19.14 + -0.84)

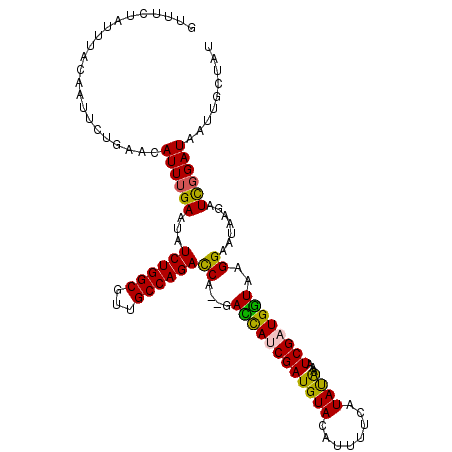
| Location | 2,303,800 – 2,303,911 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -19.78 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2303800 111 - 22224390 AUAGCAAUUAUCCGAUCUUAUUCCUUAUCAUCGAUUUUAUAUGAAAAUGUACAUCGAUGAUC--UGGUCUGGCAACGCCAGAUAUUCAAAUGUUCAGAAUUGUAAAUAAUAAC ...((((((.............((..(((((((((..(((((....))))).))))))))).--.))((((((...))))))...............)))))).......... ( -28.40) >DroSec_CAF1 1787 110 - 1 AUAGCAAUUAUCCGAUCUUAUUCCUUACCAUCGAU-UUAUAUGAAAAUGUACAUCGGUGGUC--UGGUCUGGCAACGCCAGAUAUUCAAAUGUUCAGAAUUGUAAACAGAAAC ...((((((.............((..(((((((((-.(((((....))))).))))))))).--.))((((((...))))))...............)))))).......... ( -31.80) >DroSim_CAF1 2590 110 - 1 AUAGCAAUUAUCCGAUCUUAUUCCUUACCAUCGAU-UUAUAUGAAAAUCUACAUCGGUGGUC--UGGUCUGGCAACGCCAGAUAUUCAAAUGUUCAGAAUUGUAAACAGAAAC ...((((((.............((..(((((((((-.((.((....)).)).))))))))).--.))((((((...))))))...............)))))).......... ( -27.70) >DroEre_CAF1 2932 113 - 1 AAGGAAAUUAUCCGAUUUUUUUCCUUACCAUCGAUUUCGUAUAAAAAUGUACAUCGAUGGUAGUUGAUCUGGCAACGCCAGAGAUUCAAAUGUUAAGAAUUGUAAAUAGAAAC ..(((.....)))(((((((..(.(((((((((((...(((((....))))))))))))))))((((((((((...))))))...))))..)..)))))))............ ( -30.10) >DroYak_CAF1 2638 113 - 1 AAUGAAGUGAUCAAAUUUUACUCCUUACCAUCGAUUUCGUAUUAAAAUGUACAUCGAUAAUCGUUGAUCUGGCAACGCCAGAGAUUCAAAUGCUAAGAAUUACAAAUAGAAAC ((((((((((.......))))).......((((((...((((......))))))))))..)))))..((((((...))))))(((((.........)))))............ ( -20.00) >consensus AUAGCAAUUAUCCGAUCUUAUUCCUUACCAUCGAUUUUAUAUGAAAAUGUACAUCGAUGGUC__UGGUCUGGCAACGCCAGAUAUUCAAAUGUUCAGAAUUGUAAAUAGAAAC ...((((((.................(((((((((..(((......)))...))))))))).....(((((((...)))))))..............)))))).......... (-19.78 = -20.18 + 0.40)

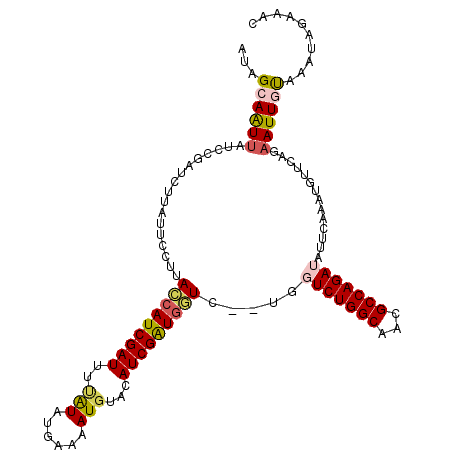
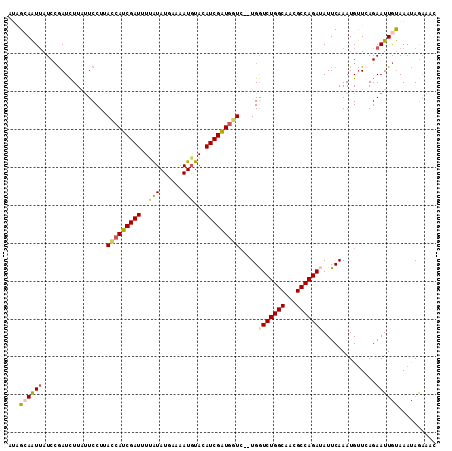
| Location | 2,303,833 – 2,303,943 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.21 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -15.34 |
| Energy contribution | -14.46 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2303833 110 - 22224390 CGUCAGUUGCUUAGUGUUCAUGAGCUUUCUGAAUAGCAAUUAUCCGAUCUUAUUCCUUAUCAUCGAUUUUAUAUGAAAAUGUACAUCGAUGAUC--UGGUCUGGCAACGCCA .((((((((((....(((((.(......)))))))))))...............((..(((((((((..(((((....))))).))))))))).--.)).)))))....... ( -28.80) >DroSec_CAF1 1820 109 - 1 CACCAGUAAUUUAGUGCUCAUGAUCAUUCUGAAUAGCAAUUAUCCGAUCUUAUUCCUUACCAUCGAU-UUAUAUGAAAAUGUACAUCGGUGGUC--UGGUCUGGCAACGCCA .(((((........((((((.((....)))))...)))....................(((((((((-.(((((....))))).))))))))))--))))..(((...))). ( -28.00) >DroSim_CAF1 2623 109 - 1 CACCAGUAAUUUAGUGCUCAUGAUCAUUCUGAAUAGCAAUUAUCCGAUCUUAUUCCUUACCAUCGAU-UUAUAUGAAAAUCUACAUCGGUGGUC--UGGUCUGGCAACGCCA .(((((........((((((.((....)))))...)))....................(((((((((-.((.((....)).)).))))))))))--))))..(((...))). ( -23.90) >DroEre_CAF1 2965 112 - 1 CAUCAAUAUUUUAGUUUUUAUGAGCGUUCUGAAAGGAAAUUAUCCGAUUUUUUUCCUUACCAUCGAUUUCGUAUAAAAAUGUACAUCGAUGGUAGUUGAUCUGGCAACGCCA .((((((......((((....)))).......(((((((..(......)..)))))))(((((((((...(((((....)))))))))))))).))))))..(((...))). ( -27.70) >DroYak_CAF1 2671 112 - 1 CAUCAAUAUUUUUGUUUUCACAAGUGUUCUGAAAUGAAGUGAUCAAAUUUUACUCCUUACCAUCGAUUUCGUAUUAAAAUGUACAUCGAUAAUCGUUGAUCUGGCAACGCCA .((((((...((((...((((.....(((......))))))).))))..............((((((...((((......))))))))))....))))))..(((...))). ( -17.20) >consensus CAUCAGUAAUUUAGUGUUCAUGAGCAUUCUGAAUAGCAAUUAUCCGAUCUUAUUCCUUACCAUCGAUUUUAUAUGAAAAUGUACAUCGAUGGUC__UGGUCUGGCAACGCCA .((((.........((((((.((....)))))))).......................(((((((((..(((......)))...)))))))))...))))..(((...))). (-15.34 = -14.46 + -0.88)

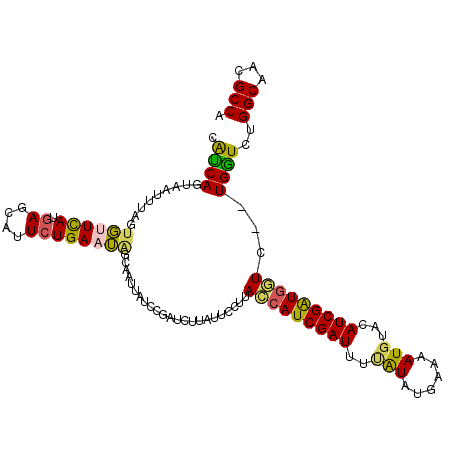
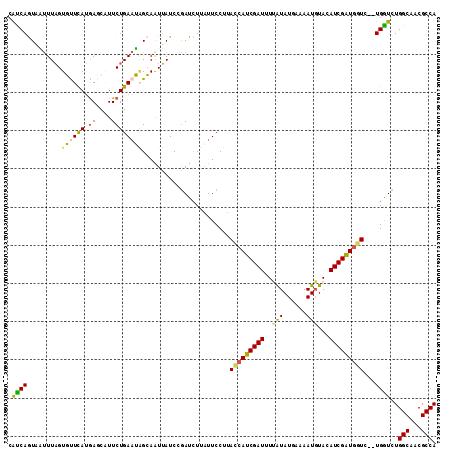
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:39 2006