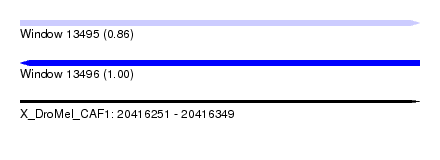
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,416,251 – 20,416,349 |
| Length | 98 |
| Max. P | 0.999027 |

| Location | 20,416,251 – 20,416,349 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.855241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20416251 98 + 22224390 -------------AAUGGGAAUGGGGAUGGGGAUGGGAUGCAGAUUCACAUCCCAGCGGAACGCAAUUUGCGUUGCACUUUUCAAUUUCCGGCAAAGGACUUCAUCACCAC -------------..(((..((((((.((..(((((((((........)))))))..(.(((((.....))))).)..))..))...(((......)))))))))..))). ( -34.60) >DroSec_CAF1 4997 99 + 1 ------------UUAUGGGACUGGGAAUGGGGAUGGGAUGCAGAUUCACAUCCCAGCGGAACGCAAUUUGCGUUGCACUUUUCAAUUUCCGGCAAAGGACUUCAUCACCAC ------------...((((.(((((((((..(((((((((........)))))))..(.(((((.....))))).)..))..).)))))))))....((.....)).))). ( -31.60) >DroSim_CAF1 2030 111 + 1 GAGUGGCGCUGGUUAUGGGACUGGGAAUGGGGAUGGGAUGCAGAUUCACAUCCCAGCGGAACGCAAUUUGCGUUGCACUUUUCAAUUUCCGGCAAAGGACUUCAUCACCAC ..((((.(..((((....(.(((((((((..(((((((((........)))))))..(.(((((.....))))).)..))..).)))))))))....))))....).)))) ( -38.40) >DroEre_CAF1 4900 98 + 1 -------------AAUUGGGAUGAGAAUGGGGAUAGGAUGCAGAUUCACAUCCCCGCGGAACGCAAUUUGCAUUGCACUUUUCAAUUUCCGGCAAAGGACUUCAUCACCAC -------------...(((((((((((.((((((..((.......))..)))))).......(((((....)))))....)))....(((......)))..))))).))). ( -28.30) >DroYak_CAF1 1616 98 + 1 -------------AAUGGGAAUGGAAAUGGAAAUGGGAUGCAGAUUCACAUCCCAGCGGAACGCAAUUUGCGUUGCACUUUUCAAUUUCCGGCAAAGGACUUCAUCACCAC -------------..(((..(((((..(((((((((((((........)))))))..(.(((((.....))))).)..))))))...(((......))).)))))..))). ( -31.90) >consensus _____________AAUGGGAAUGGGAAUGGGGAUGGGAUGCAGAUUCACAUCCCAGCGGAACGCAAUUUGCGUUGCACUUUUCAAUUUCCGGCAAAGGACUUCAUCACCAC ...............(((((.((((..(((((((((((((........)))))))..(.(((((.....))))).)..))))))...(((......))).)))))).))). (-25.20 = -25.20 + -0.00)

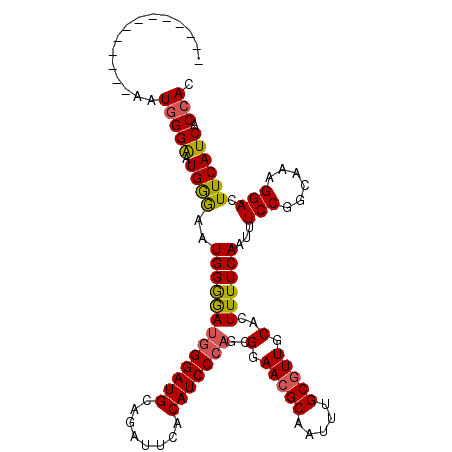
| Location | 20,416,251 – 20,416,349 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -28.84 |
| Energy contribution | -28.40 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20416251 98 - 22224390 GUGGUGAUGAAGUCCUUUGCCGGAAAUUGAAAAGUGCAACGCAAAUUGCGUUCCGCUGGGAUGUGAAUCUGCAUCCCAUCCCCAUCCCCAUUCCCAUU------------- ((((.((((...(((......)))........((((.(((((.....))))).))))(((((((......))))))).....)))).)))).......------------- ( -32.30) >DroSec_CAF1 4997 99 - 1 GUGGUGAUGAAGUCCUUUGCCGGAAAUUGAAAAGUGCAACGCAAAUUGCGUUCCGCUGGGAUGUGAAUCUGCAUCCCAUCCCCAUUCCCAGUCCCAUAA------------ .(((..(((...(((......)))........((((.(((((.....))))).))))(((((((......))))))).....)))..))).........------------ ( -30.80) >DroSim_CAF1 2030 111 - 1 GUGGUGAUGAAGUCCUUUGCCGGAAAUUGAAAAGUGCAACGCAAAUUGCGUUCCGCUGGGAUGUGAAUCUGCAUCCCAUCCCCAUUCCCAGUCCCAUAACCAGCGCCACUC ((((((.((...(((......))).((((...((((.(((((.....))))).))))(((((((......)))))))...........))))........)).)))))).. ( -35.70) >DroEre_CAF1 4900 98 - 1 GUGGUGAUGAAGUCCUUUGCCGGAAAUUGAAAAGUGCAAUGCAAAUUGCGUUCCGCGGGGAUGUGAAUCUGCAUCCUAUCCCCAUUCUCAUCCCAAUU------------- .(((.(((((........((.(((((((....)))(((((....))))).))))))(((((((.((.......)).)))))))....))))))))...------------- ( -32.30) >DroYak_CAF1 1616 98 - 1 GUGGUGAUGAAGUCCUUUGCCGGAAAUUGAAAAGUGCAACGCAAAUUGCGUUCCGCUGGGAUGUGAAUCUGCAUCCCAUUUCCAUUUCCAUUCCCAUU------------- ..((..(.(.....).)..))((((((.((((((((.(((((.....))))).))))(((((((......))))))).)))).)))))).........------------- ( -35.60) >consensus GUGGUGAUGAAGUCCUUUGCCGGAAAUUGAAAAGUGCAACGCAAAUUGCGUUCCGCUGGGAUGUGAAUCUGCAUCCCAUCCCCAUUCCCAUUCCCAUU_____________ .(((.((((...(((......)))........((((.(((((.....))))).))))(((((((......))))))).....)))).)))..................... (-28.84 = -28.40 + -0.44)

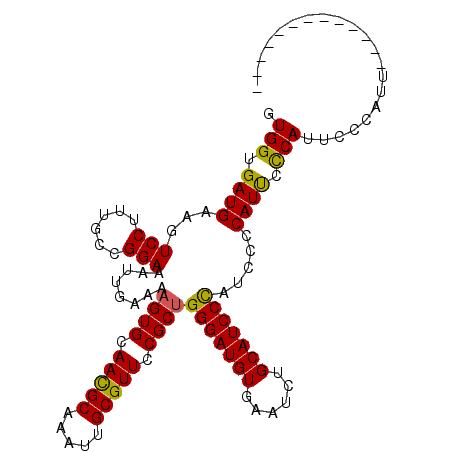

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:34 2006