
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,399,940 – 20,400,036 |
| Length | 96 |
| Max. P | 0.737292 |

| Location | 20,399,940 – 20,400,036 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -23.24 |
| Consensus MFE | -11.68 |
| Energy contribution | -11.32 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
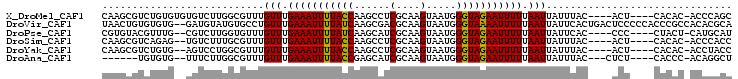
>X_DroMel_CAF1 20399940 96 + 22224390 CAAGCGUCUGUGUGUGUCUUGGCGUUUGUUUGAAAUUUUACCAAGCCUCGCAAGUAAUGGGUAGAAUUUUUAAUUAUUUAC----ACU----CACAC-ACCCAGC ...((....(((((((...((......(((.(((((((((((......(....).....))))))))))).)))......)----)..----)))))-))...)) ( -25.80) >DroVir_CAF1 72118 103 + 1 UAACUGUGUGUG--GAUGUAUGUGCCUGUUUGAAAUUUUAUCAAGCGACGCAAGUAAUGGGUAAAAUUUUUAAUUAUUCACUGACUCCCCCACCCGCCACACGCA .....(((((((--(..((..(((...(((.(((((((((((.....((....))....))))))))))).)))....)))..))...........)))))))). ( -27.52) >DroPse_CAF1 72923 94 + 1 CGUGUACGUUUG--CGUCUUGGUGUUUGUUUGAAAUUUUAUCAAGCAUCGCAAGUAAUGGGUAGAAUUUUUAAUUAUUCAC----CCC----CUACU-CAUGCAU .....(((....--)))...((((((((..(((....))).))))))))(((((((..((((.((((........))))))----)).----.))))-..))).. ( -22.90) >DroSim_CAF1 46514 94 + 1 CAAGCGUCAGAG--UGUCUUUGCGUUUGUUUGAAAUUUUACCAAGCCUCGCAAGUAAUGGGUAGAAUUUUUAAUUAUUUAC----ACU----CACAC-ACCCACC ((((((.(((((--...)))))))))))...(((((((((((......(....).....)))))))))))...........----...----.....-....... ( -21.00) >DroYak_CAF1 38588 94 + 1 CAAGCGUCUGUG--AGUCCUGGCGUUUGUUUGAAAUUUUACCAAGCCUCGCAAGUAAUGGGUAGAAUUUUUAAUUAUUUAC----ACU----CACAC-ACCUACC ..((.((.((((--(((..........(((.(((((((((((......(....).....))))))))))).))).......----)))----)))).-))))... ( -23.13) >DroAna_CAF1 74393 89 + 1 ------UGUGUG--UUUCUUGGCGUUUGUUUGAAAUUUUACCGAGCAUCGCAAGUAAUGGGUAGAAUUUUUAAUUAUUUAC---CUCU----CACCC-ACAGGCU ------((((.(--(.....((.....(((.(((((((((((.(....(....)...).))))))))))).)))......)---)...----.)).)-))).... ( -19.10) >consensus CAAGCGUCUGUG__UGUCUUGGCGUUUGUUUGAAAUUUUACCAAGCCUCGCAAGUAAUGGGUAGAAUUUUUAAUUAUUUAC____ACU____CACAC_ACCCACC ...........................(((.(((((((((((......(....).....))))))))))).)))............................... (-11.68 = -11.32 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:26 2006