
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,385,957 – 20,386,059 |
| Length | 102 |
| Max. P | 0.957568 |

| Location | 20,385,957 – 20,386,059 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.50 |
| Mean single sequence MFE | -16.92 |
| Consensus MFE | -12.65 |
| Energy contribution | -13.04 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
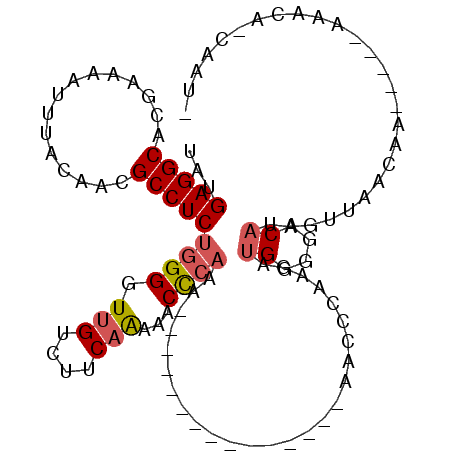
>X_DroMel_CAF1 20385957 102 - 22224390 UAUGAGGCACGAAAAUUUACAACGCCUCUGGGGUCGUCUUCAAAAACCCAAAAAAAG--AGAAAAAAAAACCAAGAUGGGGACAUAGUUAACAA-----AAACAGCAAU- ...(((((...............)))))((((..............)))).......--.........(((....(((....))).))).....-----..........- ( -15.30) >DroSec_CAF1 68334 89 - 1 UAUGAGGCACGAAAAUUUACAACGCCUCUGGGGUUGUCUUCAAAAACCCAAA---------------AACCCAAGAUGGGGACAUAGUUAACAA-----AAACAGCAAU- ...(((((...............)))))((((.(((....)))...))))..---------------..(((.....)))......(((.....-----.)))......- ( -18.46) >DroSim_CAF1 38079 89 - 1 UAUGAGGCACGAAAAUUUACAACGCCUCUGGGGUUGUCUUCAAAAACCCAAA---------------AACCCAAGAUGGGGACAUAGUUAACAA-----AAACAGCAAU- ...(((((...............)))))((((.(((....)))...))))..---------------..(((.....)))......(((.....-----.)))......- ( -18.46) >DroEre_CAF1 34189 101 - 1 UAUGAGGCACGAAAAUUUACAACGCCUCUGGGGUUGUCUUCAAAAACCCAAAAACC--------CAAAGCCCAAGAUGGGGACAAAGUUAACAACAAAAAAAGA-AAUGG ...(((((...............)))))((((.(((....)))...)))).....(--------((...(((.....)))......((.....)).........-..))) ( -18.56) >DroYak_CAF1 24389 81 - 1 UAUGAGGCACGAAAAUUUACAACGCCUCUGGGGUUGUCUUCAAAAACU-----------------------CAAGAUGGGGACAAAGUUAACAA-----AAAAA-AAUUG ...(((((...............)))))(((..(((((((((......-----------------------.....)))))))))..)))....-----.....-..... ( -15.56) >DroPer_CAF1 55043 84 - 1 UAUGAGGCACGAAAAUUUACAACGCCUCUGGGGUUGUCUUCAG--ACCCAAGACCAGCCAAAAAAAAAAACAACGAUGAAGACU------------------------GU ...(((((...............)))))...(((((((((...--....)))).))))).........................------------------------.. ( -15.16) >consensus UAUGAGGCACGAAAAUUUACAACGCCUCUGGGGUUGUCUUCAAAAACCCAAA_______________AACCCAAGAUGGGGACAUAGUUAACAA_____AAACA_CAAU_ ...(((((...............)))))((((.(((....)))...))))..........................((....)).......................... (-12.65 = -13.04 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:22 2006