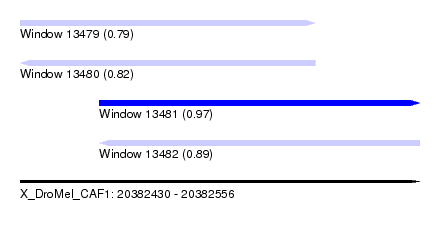
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,382,430 – 20,382,556 |
| Length | 126 |
| Max. P | 0.968943 |

| Location | 20,382,430 – 20,382,523 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -23.16 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
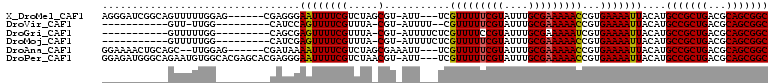
>X_DroMel_CAF1 20382430 93 + 22224390 ---------------CGAAAGGCAGGUCAUGAGUGACGGCGCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGA---AAU-ACGCUAGACG ---------------.(((((((..((((....)))).))(((((((...)))))))......)))))...((((((((......))))))))..---...-.......... ( -29.80) >DroVir_CAF1 52191 108 + 1 CGGCCCCCAAAAACCGAGCAGCCUCAUCAUGAGUGACGGCGCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACG--AAAAU-ACG-UAAACG (((..........)))....(((((((.....)))).)))(((((((...)))))))(((((.(((((...((((((((......)))))))))--)))))-)))-)..... ( -34.10) >DroGri_CAF1 46404 105 + 1 UG-----AAAUAACCGAGCAGCCCCAUCAUGAGUGACGGCGCCGCUGCGUCAGCGGCAUGUAAUUUUCACGAUUUUUCGCAAAUACGGAAAACGAGAAAAU-ACG-UAAACG ..-----.............(((...(((....))).)))(((((((...)))))))(((((.(((((.((.(((((((......))))))))).))))))-)))-)..... ( -30.90) >DroMoj_CAF1 62373 110 + 1 CGUACCCAAAAAACCGAGCAGCCUCAUCAUGAGUGACGGCGCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGAGAAAAU-ACG-UAAACG (((.................(((((((.....)))).)))(((((((...)))))))(((((.(((((.((.(((((((......))))))))).))))))-)))-)..))) ( -34.20) >DroAna_CAF1 56441 94 + 1 ---------------AGAAAGGCAGGUCAUGAGUGACGGCGCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGA---AAUUUCGCUAGACG ---------------.(((((((..((((....)))).))(((((((...)))))))......)))))...((((((((......))))))))..---.............. ( -29.70) >DroPer_CAF1 51884 93 + 1 ---------------GGGAAGGCAGGUCAUGAGUGACGGCGCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGA---AAU-ACGUUAGACG ---------------(..((.((..((((....)))).))(((((((...)))))))......))..)...((((((((......))))))))..---...-.......... ( -29.10) >consensus _______________GAGAAGCCACAUCAUGAGUGACGGCGCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGA___AAU_ACG_UAAACG .............................((((..(((..(((((((...))))))).)))....))))..((((((((......))))))))................... (-23.16 = -23.18 + 0.03)


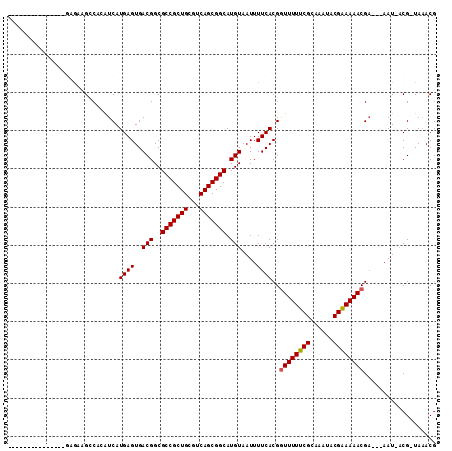
| Location | 20,382,430 – 20,382,523 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.44 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20382430 93 - 22224390 CGUCUAGCGU-AUU---UCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGCGCCGUCACUCAUGACCUGCCUUUCG--------------- ......((((-(((---(((((((((((....)))))))))...))))..)))..(((((((...))))))))).((((....))))..........--------------- ( -27.50) >DroVir_CAF1 52191 108 - 1 CGUUUA-CGU-AUUUU--CGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGCGCCGUCACUCAUGAUGAGGCUGCUCGGUUUUUGGGGGCCG ......-.((-(((((--((((((((((....)))))))))...))))).)))..(((((((...)))))))..(((((....))))).((((.((........)).)))). ( -36.80) >DroGri_CAF1 46404 105 - 1 CGUUUA-CGU-AUUUUCUCGUUUUCCGUAUUUGCGAAAAAUCGUGAAAAUUACAUGCCGCUGACGCAGCGGCGCCGUCACUCAUGAUGGGGCUGCUCGGUUAUUU-----CA ......-.((-((((((.((((((.(((....))).)))).)).))))).)))..(((((....))((((((.((((((....)))))).)))))).))).....-----.. ( -33.70) >DroMoj_CAF1 62373 110 - 1 CGUUUA-CGU-AUUUUCUCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGCGCCGUCACUCAUGAUGAGGCUGCUCGGUUUUUUGGGUACG ......-.((-((((((.((((((((((....)))))))).)).))))).)))..(((((((...)))))))(((.((((....).))))))(((((((....))))))).. ( -37.90) >DroAna_CAF1 56441 94 - 1 CGUCUAGCGAAAUU---UCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGCGCCGUCACUCAUGACCUGCCUUUCU--------------- ((((.((((.....---..(((((((((....)))))))))((((.......)))).))))))))....((((..((((....)))).)))).....--------------- ( -27.80) >DroPer_CAF1 51884 93 - 1 CGUCUAACGU-AUU---UCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGCGCCGUCACUCAUGACCUGCCUUCCC--------------- ........((-(((---(((((((((((....)))))))))...))))..)))..(((((((...)))))))((.((((....))))..))......--------------- ( -26.90) >consensus CGUCUA_CGU_AUU___UCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGCGCCGUCACUCAUGACCUGCCUGCCC_______________ ...................(((((((((....)))))))))(((((........((((((((...)))))))).......)))))........................... (-24.42 = -24.44 + 0.03)

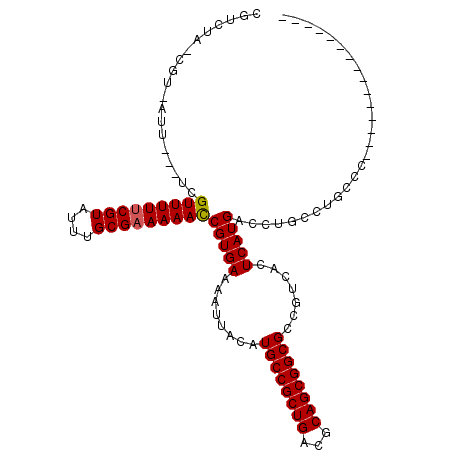
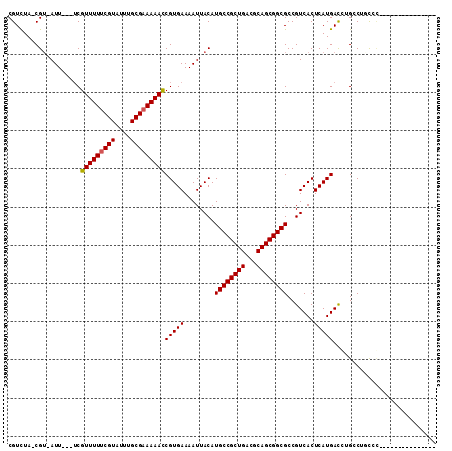
| Location | 20,382,455 – 20,382,556 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -19.85 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20382455 101 + 22224390 GCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGA---AAU-ACGCUAGACGAAAAUUCCCUCG------CUCCAAAAACUGCCGAUCCCU (((((((...)))))))....(((((((...((((((((......))))))))..---...-.(....)..)))))))...(((------(..........).)))..... ( -23.30) >DroVir_CAF1 52231 86 + 1 GCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACG--AAAAU-ACG-UAAACGAAAACUGGAUG---------CCAA-AAC----------- (((((((...)))))))(((((.(((((...((((((((......)))))))))--)))))-)))-)..........(((...---------))).-...----------- ( -25.10) >DroGri_CAF1 46439 89 + 1 GCCGCUGCGUCAGCGGCAUGUAAUUUUCACGAUUUUUCGCAAAUACGGAAAACGAGAAAAU-ACG-UAAACGAAAACUCGCUG---------CCAAAAAC----------- (((((((...)))))))(((((.(((((.((.(((((((......))))))))).))))))-)))-)...(((....)))...---------........----------- ( -26.50) >DroMoj_CAF1 62413 89 + 1 GCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGAGAAAAU-ACG-UAAACGAAAACUCGAUG---------CCAAAAAC----------- (((((((...)))))))(((((.(((((.((.(((((((......))))))))).))))))-)))-)...(((....)))...---------........----------- ( -27.70) >DroAna_CAF1 56466 100 + 1 GCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGA---AAUUUCGCUAGACGAAAAUUUUAUCG------CUCCAA--GCUGCAGUUUUCC (((((((...))))))).((((.........((((((((......))))))))..---......(((.(((((........)))------.))..)--))))))....... ( -27.10) >DroPer_CAF1 51909 107 + 1 GCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGA---AAU-ACGUUAGACGAAAAUUCCCUCGUGCUCGUGCCACAUUCUGCCCAUCUCC (((((((...)))))))..........((((((((((((......))))))))..---...-.......((((........))))...))))................... ( -27.20) >consensus GCCGCUGCGUCAGCGGCAUGUAAUUUUCACGGUUUUUCGCAAAUACGAAAAACGA___AAU_ACG_UAAACGAAAACUCGAUC_________CCAAAAAC___________ (((((((...)))))))..............((((((((......)))))))).......................................................... (-19.85 = -19.88 + 0.03)



| Location | 20,382,455 – 20,382,556 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -22.00 |
| Energy contribution | -21.78 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20382455 101 - 22224390 AGGGAUCGGCAGUUUUUGGAG------CGAGGGAAUUUUCGUCUAGCGU-AUU---UCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGC .......((((((..((...(------((..(((((...((.....)).-)))---))(((((((((....))))))))))))...))..)).))))((((...))))... ( -28.40) >DroVir_CAF1 52231 86 - 1 -----------GUU-UUGG---------CAUCCAGUUUUCGUUUA-CGU-AUUUU--CGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGC -----------...-..((---------(((..((((((((....-)..-.....--.(((((((((....)))))))))...)))))))..)))))((((...))))... ( -24.30) >DroGri_CAF1 46439 89 - 1 -----------GUUUUUGG---------CAGCGAGUUUUCGUUUA-CGU-AUUUUCUCGUUUUCCGUAUUUGCGAAAAAUCGUGAAAAUUACAUGCCGCUGACGCAGCGGC -----------((..((..---------((.(((.(((((((.((-((.-..............))))...))))))).)))))..))..))..(((((((...))))))) ( -26.16) >DroMoj_CAF1 62413 89 - 1 -----------GUUUUUGG---------CAUCGAGUUUUCGUUUA-CGU-AUUUUCUCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGC -----------..((((.(---------(...(((....((....-)).-.....)))(((((((((....))))))))).)).))))......(((((((...))))))) ( -26.50) >DroAna_CAF1 56466 100 - 1 GGAAAACUGCAGC--UUGGAG------CGAUAAAAUUUUCGUCUAGCGAAAUU---UCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGC .((((......((--(.(((.------(((........)))))))))....))---))(((((((((....))))))))).((((...))))..(((((((...))))))) ( -29.10) >DroPer_CAF1 51909 107 - 1 GGAGAUGGGCAGAAUGUGGCACGAGCACGAGGGAAUUUUCGUCUAACGU-AUU---UCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGC .............((((((......((((..(((((...((.....)).-)))---))(((((((((....)))))))))))))....))))))(((((((...))))))) ( -31.10) >consensus ___________GUUUUUGG_________CAGCGAAUUUUCGUCUA_CGU_AUU___UCGUUUUUCGUAUUUGCGAAAAACCGUGAAAAUUACAUGCCGCUGACGCAGCGGC .................................((((((((.....)...........(((((((((....)))))))))...)))))))....(((((((...))))))) (-22.00 = -21.78 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:21 2006