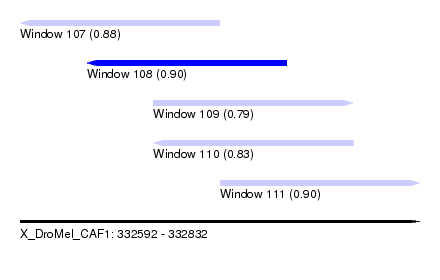
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 332,592 – 332,832 |
| Length | 240 |
| Max. P | 0.903066 |

| Location | 332,592 – 332,712 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -37.39 |
| Energy contribution | -37.33 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 332592 120 - 22224390 UGCAUGGAACUGACAAAAACACGGUUUCAACUUGGCAUCAACCUGCUUAAGAUCACCUCCGAGGAGGUGGUAAGCAACAGGGGCGACCUGACCACCGAAUGGAAGCAGUACUUCUACUCC (((.((((((((.........)))))))).(((((((......))).))))((((((((....))))))))..))).((((.....))))......((.((((((.....)))))).)). ( -38.40) >DroSec_CAF1 33 120 - 1 UGCAUGGAACUGACAAAAACACGGUUUCAGCUUGGCAUCAACCUGCUUAAGAUCACCUCCGAGGAGGUGGUAAGCAACCGGGGCGACCUGACCACCGAAUGGAAGCAGUACUUCUACUCC .((.((((((((.........)))))))))).(((........(((((...((((((((....)))))))))))))..((((....)))).)))..((.((((((.....)))))).)). ( -40.30) >DroEre_CAF1 41 120 - 1 UGCAUGGAACUGACAAAAACACGGUUUCAGCUUGGCAUCAACCUGCUUAGGAUCACCUCUGAGGAGGUGGUAAGCAACAGGGGCGACCUGACCACCGAAUGGAAGCAGUACUUCUACUCC .((.((((((((.........)))))))))).(((........(((((...((((((((....))))))))))))).((((.....)))).)))..((.((((((.....)))))).)). ( -38.60) >DroYak_CAF1 2 120 - 1 UGCAUGGAACUGACGAAAACACGGUUUCAGCUCGGCAUCAACCUGCUCAAGAUCACCUCCGAGGAGGUGGUGAGCAACAGGGGCGACCUGACCACCGAAUGGAAGCAGUACUUCUACUCC .((.((((((((.........))))))))))((((........((((((...(((((((....))))))))))))).((((.....))))....)))).((((((.....)))))).... ( -43.30) >consensus UGCAUGGAACUGACAAAAACACGGUUUCAGCUUGGCAUCAACCUGCUUAAGAUCACCUCCGAGGAGGUGGUAAGCAACAGGGGCGACCUGACCACCGAAUGGAAGCAGUACUUCUACUCC .((.((((((((.........))))))))))((((........(((((...((((((((....))))))))))))).((((.....))))....)))).((((((.....)))))).... (-37.39 = -37.33 + -0.06)

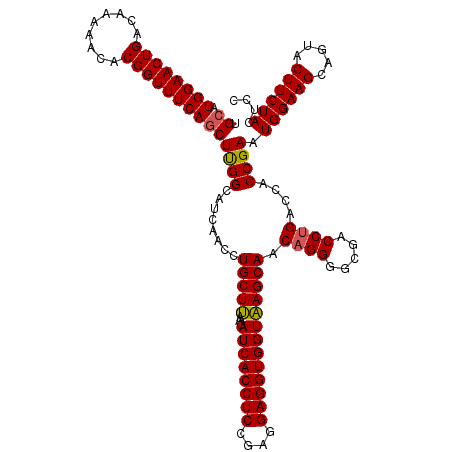
| Location | 332,632 – 332,752 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -38.15 |
| Consensus MFE | -33.72 |
| Energy contribution | -34.10 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 332632 120 - 22224390 GAGGGAGUUCCCUGAACAGGACCCCAAUUACAUGCAACACUGCAUGGAACUGACAAAAACACGGUUUCAACUUGGCAUCAACCUGCUUAAGAUCACCUCCGAGGAGGUGGUAAGCAACAG .((((....))))......((..((((...((((((....))))))((((((.........))))))....))))..))....(((((...((((((((....))))))))))))).... ( -37.30) >DroSec_CAF1 73 120 - 1 GAGGGAGUUCCCUGAACAGGACCCCAAUUACAUGCAACACUGCAUGGAACUGACAAAAACACGGUUUCAGCUUGGCAUCAACCUGCUUAAGAUCACCUCCGAGGAGGUGGUAAGCAACCG ..(..((((((((....)))..........((((((....)))))))))))..).......(((((...((((((((......))))....((((((((....))))))))))))))))) ( -39.10) >DroEre_CAF1 81 120 - 1 GAGGGAGUUCCCCGAACAGGACCCAAACUACAUGCAGCACUGCAUGGAACUGACAAAAACACGGUUUCAGCUUGGCAUCAACCUGCUUAGGAUCACCUCUGAGGAGGUGGUAAGCAACAG ..(((.....)))......((.((((.((.((((((....))))))((((((.........)))))).)).))))..))....(((((...((((((((....))))))))))))).... ( -35.40) >DroYak_CAF1 42 120 - 1 GCGCGAGUUCCCCGAACAGGACCCCAACUACAUGCAGCACUGCAUGGAACUGACGAAAACACGGUUUCAGCUCGGCAUCAACCUGCUCAAGAUCACCUCCGAGGAGGUGGUGAGCAACAG ((.((((((..((.....))..........((((((....))))))((((((.........)))))).)))))))).......((((((...(((((((....))))))))))))).... ( -40.80) >consensus GAGGGAGUUCCCCGAACAGGACCCCAACUACAUGCAACACUGCAUGGAACUGACAAAAACACGGUUUCAGCUUGGCAUCAACCUGCUUAAGAUCACCUCCGAGGAGGUGGUAAGCAACAG ..(((.((((........))))))).....((((((....))))))((((((.........))))))..((((((((......))))....((((((((....))))))))))))..... (-33.72 = -34.10 + 0.38)

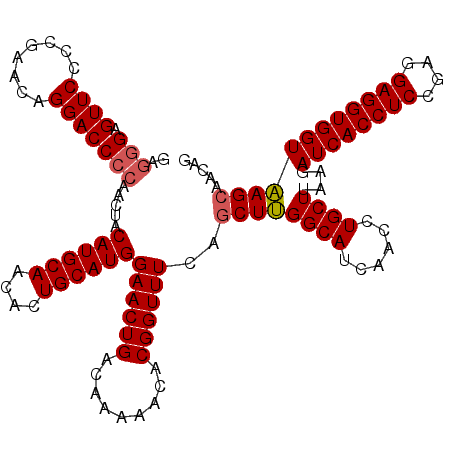
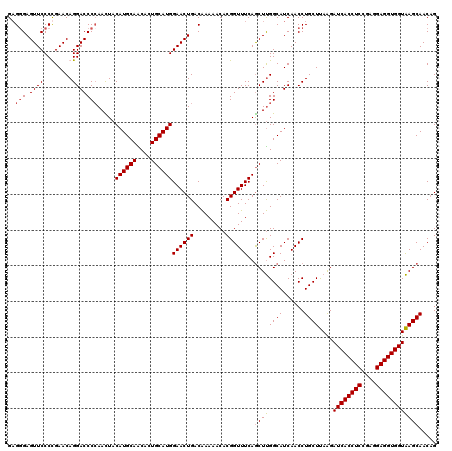
| Location | 332,672 – 332,792 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -43.33 |
| Consensus MFE | -39.91 |
| Energy contribution | -40.60 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 332672 120 + 22224390 UGAUGCCAAGUUGAAACCGUGUUUUUGUCAGUUCCAUGCAGUGUUGCAUGUAAUUGGGGUCCUGUUCAGGGAACUCCCUCUUUCCGAGUAGGGCUAUCAACUGGGCCAGUGUGUCCCUGU ....(((.((((((.............((((((.(((((......))))).))))))((((((((((((((....))))......)))))))))).)))))).)))(((.......))). ( -38.40) >DroSec_CAF1 113 120 + 1 UGAUGCCAAGCUGAAACCGUGUUUUUGUCAGUUCCAUGCAGUGUUGCAUGUAAUUGGGGUCCUGUUCAGGGAACUCCCUCUUUCCGAGCAGGGCUAUCAACUGGGCCAGUGUGUCCCUGU .(((((...((((...(((.(((....((((((.(((((......))))).))))))((((((((((((((....))))......))))))))))...))))))..)))))))))..... ( -41.00) >DroEre_CAF1 121 120 + 1 UGAUGCCAAGCUGAAACCGUGUUUUUGUCAGUUCCAUGCAGUGCUGCAUGUAGUUUGGGUCCUGUUCGGGGAACUCCCUCUUUCCGAGCAGGGCUAUCAACUGGGCCAACGUGUCCCUCU (((((((((((((((((...))))..........((((((....))))))))))))))(((((((((((..(........)..))))))))))))))))...(((.(.....).)))... ( -43.80) >DroYak_CAF1 82 120 + 1 UGAUGCCGAGCUGAAACCGUGUUUUCGUCAGUUCCAUGCAGUGCUGCAUGUAGUUGGGGUCCUGUUCGGGGAACUCGCGCUUUCCGAGCAGGGCUAUCAGCUGGGCCAGUGUGUCCCUGU ....(((.((((((.............(((..(.((((((....)))))).)..)))((((((((((((..(........)..)))))))))))).)))))).)))(((.......))). ( -50.10) >consensus UGAUGCCAAGCUGAAACCGUGUUUUUGUCAGUUCCAUGCAGUGCUGCAUGUAAUUGGGGUCCUGUUCAGGGAACUCCCUCUUUCCGAGCAGGGCUAUCAACUGGGCCAGUGUGUCCCUGU ....(((.((((((.............((((((.((((((....)))))).))))))((((((((((((..(........)..)))))))))))).)))))).))).............. (-39.91 = -40.60 + 0.69)

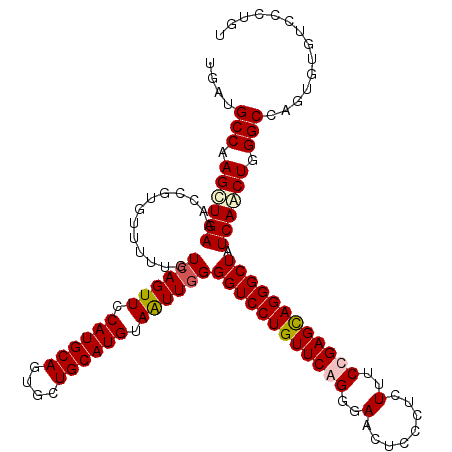
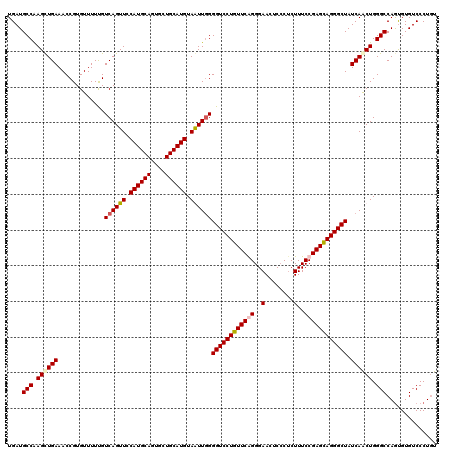
| Location | 332,672 – 332,792 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -32.41 |
| Energy contribution | -32.35 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 332672 120 - 22224390 ACAGGGACACACUGGCCCAGUUGAUAGCCCUACUCGGAAAGAGGGAGUUCCCUGAACAGGACCCCAAUUACAUGCAACACUGCAUGGAACUGACAAAAACACGGUUUCAACUUGGCAUCA .(((.......)))(((.((((((.((((...(((.....)))..((((((((....)))..........((((((....)))))))))))...........)))))))))).))).... ( -36.70) >DroSec_CAF1 113 120 - 1 ACAGGGACACACUGGCCCAGUUGAUAGCCCUGCUCGGAAAGAGGGAGUUCCCUGAACAGGACCCCAAUUACAUGCAACACUGCAUGGAACUGACAAAAACACGGUUUCAGCUUGGCAUCA .(((.......)))(((.((((((.((((.((.((((.....(((.((((........))))))).....((((((....))))))...))))......)).)))))))))).))).... ( -37.40) >DroEre_CAF1 121 120 - 1 AGAGGGACACGUUGGCCCAGUUGAUAGCCCUGCUCGGAAAGAGGGAGUUCCCCGAACAGGACCCAAACUACAUGCAGCACUGCAUGGAACUGACAAAAACACGGUUUCAGCUUGGCAUCA ..............(((.((((((.((((.(((((.....)))..(((((.((.....))..........((((((....)))))))))))........)).)))))))))).))).... ( -34.30) >DroYak_CAF1 82 120 - 1 ACAGGGACACACUGGCCCAGCUGAUAGCCCUGCUCGGAAAGCGCGAGUUCCCCGAACAGGACCCCAACUACAUGCAGCACUGCAUGGAACUGACGAAAACACGGUUUCAGCUCGGCAUCA .(((.......)))(((.((((((.(((((((.((((..(((....)))..)))).)))...........((((((....))))))................)))))))))).))).... ( -43.70) >consensus ACAGGGACACACUGGCCCAGUUGAUAGCCCUGCUCGGAAAGAGGGAGUUCCCCGAACAGGACCCCAACUACAUGCAACACUGCAUGGAACUGACAAAAACACGGUUUCAGCUUGGCAUCA .((((....).)))(((.((((((.((((...(((.....)))..(((((((......))..........((((((....)))))))))))...........)))))))))).))).... (-32.41 = -32.35 + -0.06)

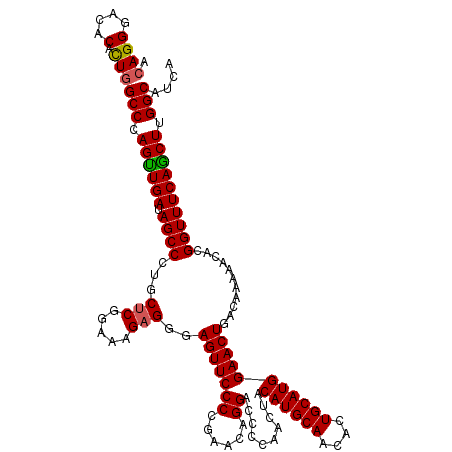
| Location | 332,712 – 332,832 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -48.65 |
| Consensus MFE | -44.19 |
| Energy contribution | -44.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 332712 120 + 22224390 GUGUUGCAUGUAAUUGGGGUCCUGUUCAGGGAACUCCCUCUUUCCGAGUAGGGCUAUCAACUGGGCCAGUGUGUCCCUGUGCCGUUUGGCCACAUUUGGAGUGGCUACCAGUUGGGCAUA ....(((..........((((((((((((((....))))......)))))))))).(((((((((((((.......))).)))...(((((((.......))))))).)))))))))).. ( -43.60) >DroSec_CAF1 153 120 + 1 GUGUUGCAUGUAAUUGGGGUCCUGUUCAGGGAACUCCCUCUUUCCGAGCAGGGCUAUCAACUGGGCCAGUGUGUCCCUGUGCCGUUUGGCCACAUUUGGAGUGGCUCCCAGUUGGGCAUA ....(((..........((((((((((((((....))))......)))))))))).(((((((((((((.......))).)))....((((((.......))))))..)))))))))).. ( -47.10) >DroEre_CAF1 161 120 + 1 GUGCUGCAUGUAGUUUGGGUCCUGUUCGGGGAACUCCCUCUUUCCGAGCAGGGCUAUCAACUGGGCCAACGUGUCCCUCUGCCGUUUGGCCACAUUUGAAGUGGCUCCCAGUUGGGCAUA (((((............((((((((((((..(........)..))))))))))))..((((((((..((((.((......)))))).((((((.......))))))))))))))))))). ( -52.80) >DroYak_CAF1 122 120 + 1 GUGCUGCAUGUAGUUGGGGUCCUGUUCGGGGAACUCGCGCUUUCCGAGCAGGGCUAUCAGCUGGGCCAGUGUGUCCCUGUGCCGUUUGGCCACAUUCGGAGUGGCUCCCAGUUGGGCGUA ..(((............((((((((((((..(........)..))))))))))))..(((((((((((.(.((....((((((....)).))))..)).).))))..))))))))))... ( -51.10) >consensus GUGCUGCAUGUAAUUGGGGUCCUGUUCAGGGAACUCCCUCUUUCCGAGCAGGGCUAUCAACUGGGCCAGUGUGUCCCUGUGCCGUUUGGCCACAUUUGGAGUGGCUCCCAGUUGGGCAUA ....(((..........((((((((((((..(........)..)))))))))))).(((((((((((((.......))).)))....((((((.......))))))..)))))))))).. (-44.19 = -44.62 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:38 2006