
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,363,757 – 20,363,862 |
| Length | 105 |
| Max. P | 0.967089 |

| Location | 20,363,757 – 20,363,862 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -13.45 |
| Energy contribution | -15.82 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
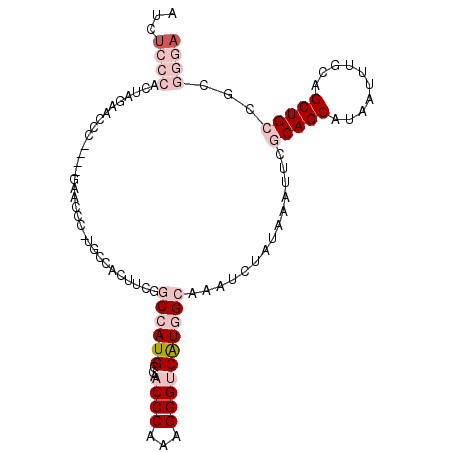
>X_DroMel_CAF1 20363757 105 + 22224390 AUCUCCCACUAGAACCCACUAGAACCCAUGCCACUUCGGCCAUGGCAACCCAAAGGGUCAUGGCAAAUCUAUAAAUUCGCAGCAUAAUUUGCAGCUGCCGCGGCA ........((((......))))......((((......((((((((..((....))))))))))..............(((((..........)))))...)))) ( -29.50) >DroPse_CAF1 32694 79 + 1 --CUCCAACUUGGUCGC------------------UGGACCAUGGCAACCCAUAGGGACAUAGAGGAUCUAUAAAUUCACAGCAUAAUUGGCAGCUGCU------ --(((((((.(((((..------------------..))))).)((..(((...)))..((((.....)))).........))....)))).)).....------ ( -16.20) >DroSec_CAF1 11141 105 + 1 AUCUCCCACUAGAACCCACUAGAACCCCUGCCACUUCGGCCAUGGAAACCCAAAGGGUCAUGGCAAAUCUAUAAAUUCGCAGCAUAAUUUGCAGCUGCCGCGGGA ........((((......))))....(((((.......((((((...((((...))))))))))..............(((((..........))))).))))). ( -31.10) >DroSim_CAF1 11589 105 + 1 AUCUCCCACUAGAACCCACUAGAACCCAUGCCACUUCGGCCAUGGCAACCCAAAGGGUCAUGGCAAAUCUAUAAAUUCGCAGCAUAAUUUGCAGCUGCCGCGGGA ...((((.((((......)))).......((.......((((((((..((....))))))))))..............(((((..........))))).)))))) ( -31.10) >DroEre_CAF1 10525 95 + 1 AUCUCCCACUA----------GAACCCCUGCCACUUUGGCCAUGGCAACCCAAAGGGUCGUGGCAAAUCUAUAAAUUCGCAGCAUAAUUUGCAGCUGCGGCGGGA ...........----------.....((((((......((((((((..((....))))))))))..............(((((..........))))))))))). ( -33.50) >DroPer_CAF1 32830 84 + 1 GUCUCUCACUUGGUCCC------------------UGGACCAUGGCAACCCAUAGGGACAUAGAGGAUCUAUAAAUUCACAGCAUAAUUGGCAGCUGCUG---GA ((((((.....((((..------------------..))))((((....))))))))))((((.....))))...((((((((..........)))).))---)) ( -23.40) >consensus AUCUCCCACUAGAACCC____GAACCC_UGCCACUUCGGCCAUGGCAACCCAAAGGGUCAUGGCAAAUCUAUAAAUUCGCAGCAUAAUUUGCAGCUGCCGCGGGA ...((((...............................((((((...((((...))))))))))..............(((((..........)))))...)))) (-13.45 = -15.82 + 2.36)

| Location | 20,363,757 – 20,363,862 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -19.06 |
| Energy contribution | -21.12 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
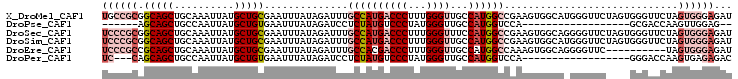
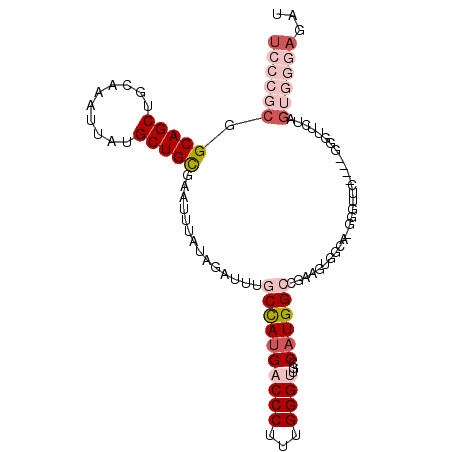
>X_DroMel_CAF1 20363757 105 - 22224390 UGCCGCGGCAGCUGCAAAUUAUGCUGCGAAUUUAUAGAUUUGCCAUGACCCUUUGGGUUGCCAUGGCCGAAGUGGCAUGGGUUCUAGUGGGUUCUAGUGGGAGAU ..((((.(((((..........)))))(((((((((((....(((((.((((((((.(......).)))))).)))))))..))).))))))))..))))..... ( -38.90) >DroPse_CAF1 32694 79 - 1 ------AGCAGCUGCCAAUUAUGCUGUGAAUUUAUAGAUCCUCUAUGUCCCUAUGGGUUGCCAUGGUCCA------------------GCGACCAAGUUGGAG-- ------.(((((..........))))).....(((((.....)))))...((((((....))))))((((------------------((......)))))).-- ( -22.90) >DroSec_CAF1 11141 105 - 1 UCCCGCGGCAGCUGCAAAUUAUGCUGCGAAUUUAUAGAUUUGCCAUGACCCUUUGGGUUUCCAUGGCCGAAGUGGCAGGGGUUCUAGUGGGUUCUAGUGGGAGAU ((((((.(((((..........)))))(((((((((((...(((.((.((((((((.(......).)))))).))))..)))))).))))))))..))))))... ( -40.30) >DroSim_CAF1 11589 105 - 1 UCCCGCGGCAGCUGCAAAUUAUGCUGCGAAUUUAUAGAUUUGCCAUGACCCUUUGGGUUGCCAUGGCCGAAGUGGCAUGGGUUCUAGUGGGUUCUAGUGGGAGAU ((((((.(((((..........)))))(((((((((((....(((((.((((((((.(......).)))))).)))))))..))).))))))))..))))))... ( -43.40) >DroEre_CAF1 10525 95 - 1 UCCCGCCGCAGCUGCAAAUUAUGCUGCGAAUUUAUAGAUUUGCCACGACCCUUUGGGUUGCCAUGGCCAAAGUGGCAGGGGUUC----------UAGUGGGAGAU ((((((((((((..........))))))......(((((((((((((((((...)))))((....))....))))))))...))----------))))))))... ( -40.00) >DroPer_CAF1 32830 84 - 1 UC---CAGCAGCUGCCAAUUAUGCUGUGAAUUUAUAGAUCCUCUAUGUCCCUAUGGGUUGCCAUGGUCCA------------------GGGACCAAGUGAGAGAC ((---(((((...........))))).))...........((((..((((((((((....)))).....)------------------)))))......)))).. ( -21.10) >consensus UCCCGCGGCAGCUGCAAAUUAUGCUGCGAAUUUAUAGAUUUGCCAUGACCCUUUGGGUUGCCAUGGCCGAAGUGGCA_GGGUUC____GGGUUCUAGUGGGAGAU ((((((.(((((..........)))))..............((((((((((...))))...)))))).............................))))))... (-19.06 = -21.12 + 2.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:10 2006