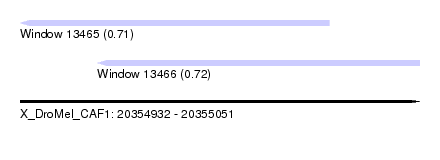
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,354,932 – 20,355,051 |
| Length | 119 |
| Max. P | 0.721928 |

| Location | 20,354,932 – 20,355,024 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 84.91 |
| Mean single sequence MFE | -19.17 |
| Consensus MFE | -14.95 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20354932 92 - 22224390 UUUUCCAAUUCCAUUUCCACGCCUG----CAAUUGCGAUUGCGAUUGCGAUUGCAGCUGCUGAUGCCGCCAUUUUAUCUAAUUGCGUUGCACCCAA .................((.(((((----((((((((((....)))))))))))))..)))).((((((.(((......))).)))..)))..... ( -24.00) >DroSec_CAF1 2340 96 - 1 UUUUCCAAUUCCAUUUCCACGCCUUUCCACGCCUGCAAUUGCGAUUGCGAUUGCAGCUGCUGAUGCCGCCAUUUUAUCUAAUUGCGUUGCACCCAA ...........................((.((((((((((((....))))))))))..)))).((((((.(((......))).)))..)))..... ( -20.80) >DroSim_CAF1 2333 90 - 1 UUUUCCAAUUCCAUUUCCACGCCUUUCCACGCCUGCAAUUGC------GAUUGCAGCUGCUGAUGCCGCCAUUUUAUCUAAUUGCGUUGCACCCAA ..............................((.((((((((.------(((....((.((....)).))......)))))))))))..))...... ( -14.30) >DroEre_CAF1 2324 83 - 1 UUUCCC------AUUUCCACGCCCCUCCACGCCUGCAGUUGC------GCUUGCAGCUGCUGAUGCCGCCAUUUUAUCUAAUUGCGUUGC-CCCAA ......------........((........((..((((((((------....))))))))....))(((.(((......))).)))..))-..... ( -17.60) >consensus UUUUCCAAUUCCAUUUCCACGCCUUUCCACGCCUGCAAUUGC______GAUUGCAGCUGCUGAUGCCGCCAUUUUAUCUAAUUGCGUUGCACCCAA ..................((((...........(((((((((....)))))))))((.((....)).))..............))))......... (-14.95 = -15.82 + 0.88)



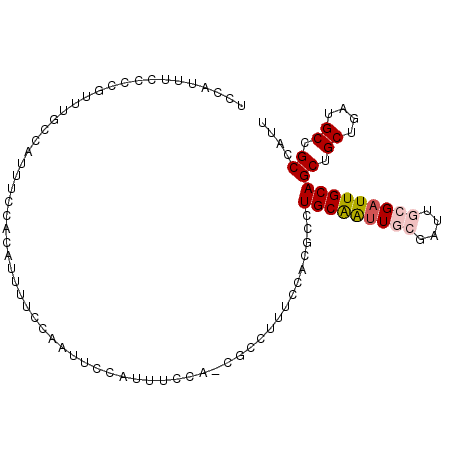
| Location | 20,354,955 – 20,355,051 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -11.82 |
| Energy contribution | -12.42 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20354955 96 - 22224390 UCCAUUUCCCCGUUUGCCAUUUCCACAUUUUCCAAUUCCAUUUCCA-CGCCUG----CAAUUGCGAUUGCGAUUGCGAUUGCAGCUGCUGAUGCCGCCAUU ......................................((((..((-.((.((----((((((((((....))))))))))))))))..))))........ ( -20.70) >DroSec_CAF1 2363 100 - 1 UCCAUUUCCCCGUUUGCCAUUUCCACAUUUUCCAAUUCCAUUUCCA-CGCCUUUCCACGCCUGCAAUUGCGAUUGCGAUUGCAGCUGCUGAUGCCGCCAUU ..........((((.((.............................-.((........))((((((((((....))))))))))..)).))))........ ( -17.10) >DroSim_CAF1 2356 94 - 1 UCCAUUUCCCCGUUUGCCAUUUCCACAUUUUCCAAUUCCAUUUCCA-CGCCUUUCCACGCCUGCAAUUGC------GAUUGCAGCUGCUGAUGCCGCCAUU ..........((((.((.............................-.((........))(((((((...------.)))))))..)).))))........ ( -10.40) >DroEre_CAF1 2346 88 - 1 UCCAUUUCCCCGUUUGCCAUUUCCACAUUUCCC------AUUUCCA-CGCCCCUCCACGCCUGCAGUUGC------GCUUGCAGCUGCUGAUGCCGCCAUU .................................------.......-...........((..((((((((------....))))))))....))....... ( -13.90) >DroAna_CAF1 18323 91 - 1 UCCAUUUCCACAGC-GGCAUGUCUGCAGU-------GCCGUUGCCGUUGCCGUUGCA--AUUGCGAUUGCGAUUGCGAUUGCAGCUGCUGAUGCCGCCAUU ............((-((((..((.(((((-------((.(((((.(((((.(((((.--...))))).))))).))))).))).)))).)))))))).... ( -40.70) >consensus UCCAUUUCCCCGUUUGCCAUUUCCACAUUUUCCAAUUCCAUUUCCA_CGCCUUUCCACGCCUGCAAUUGCGAUUGCGAUUGCAGCUGCUGAUGCCGCCAUU .............................................................(((((((((....)))))))))((.((....)).)).... (-11.82 = -12.42 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:08 2006